Figure 6.
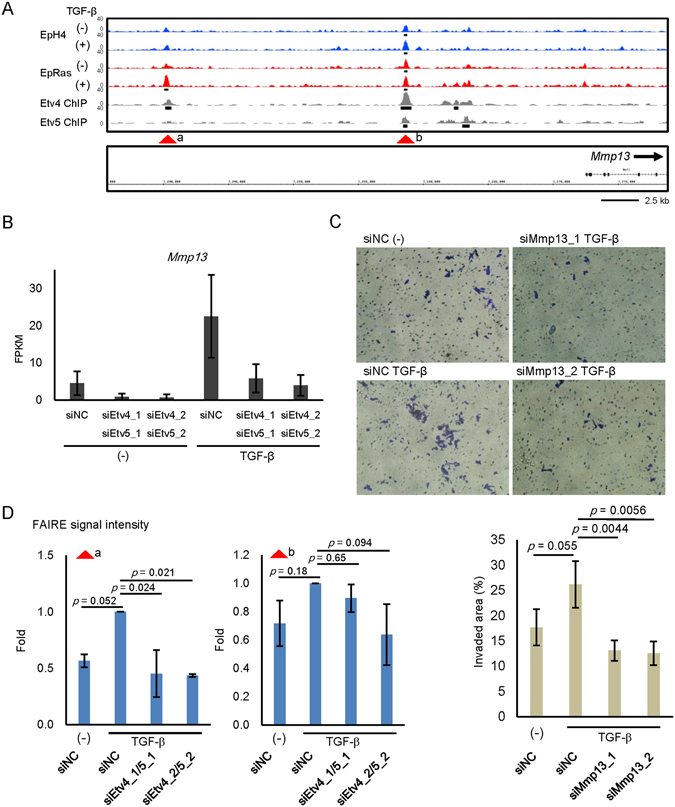
Regulation of Mmp13 expression by Etv4 and Etv5. (A) FAIRE-seq and ChIP-seq data at the matrix metalloproteinase 13 (Mmp13) gene locus as a target of Etv4 and 5. Data are shown as in Figs 2A and 4E. Arrow heads show the positions evaluated by FAIRE-qPCR in (D). (B) RNA-seq data of siEtv4/5-transfected EpRas cells treated with TGF-β. siNC; negative control siRNA. Error bars, 95% confidence intervals. (C) Matrigel invasion assay in EpRas cells. Cells were transfected with the indicated siRNAs and then seeded on the Matrigel-coated plate, incubated for 48 h with TGF-β, and fixed. (upper panels) Representative images of the cells that migrated through the Matrigel-coated membrane. (bottom graph) Quantified data representing the means of four independent experiments. Error bars, standard deviations. (D) Changes in the FAIRE-seq signal intensities by Etv4/5 siRNAs at FAIRE-positive regions of Mmp13 gene locus shown in (A). EpRas cells transfected with the indicated siRNAs were treated with TGF-β for 4 days and fixed for FAIRE-seq data acquisition. The data represent the result of two biological replicates. Error bars, standard deviations.
