Figure 1.
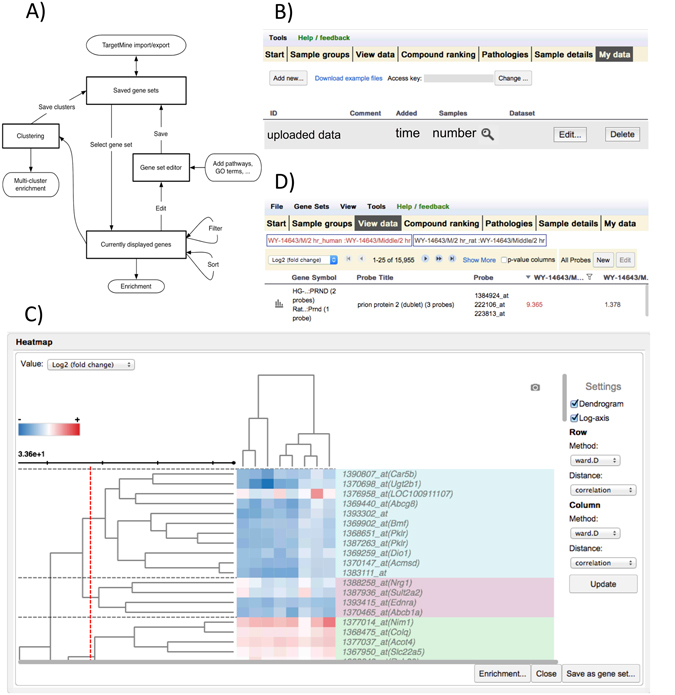
(A) Many functions in Toxygates now communicate with each other by modifying or transferring gene sets. Currently displayed genes can be filtered, sorted and tested for enrichment, and saved as a named set. They can also be clustered, and by setting a dendrogram cutoff, new gene sets can be generated, as well as enriched together. Gene sets are saved as local data in the web browser. They may be manually edited, for example by adding pathways and GO terms, and saved gene sets may be synchronised with TargetMine for further analysis and long term storage. (B) The My Data screen. Here, users can upload, adjust, and delete their own expression data as sets of samples (called batches). (C) Interactive display of heatmap with dendrogram. After selecting compounds and experimental conditions and filtering genes of interest, a clustering may be performed and displayed as a heatmap in a new pop-up window by selecting “Show heatmap” from “Tools” pull-down menu at the top of the main display. The parameters of the clustering may be changed by using the controls on the right side of the display. (D) When the currently defined sample groups contain more than one platform (i.e. more than one species), the orthologous mode is activated. This uses pre-computed groups of probes (based on amino acid sequence similarity) to display orthologous genes together as a single row. This allows for meaningful cross-species analysis, which is especially useful since Open TG-GATEs contains both rat and human data. The high-resolution image file is available at Scientific Reports online.
