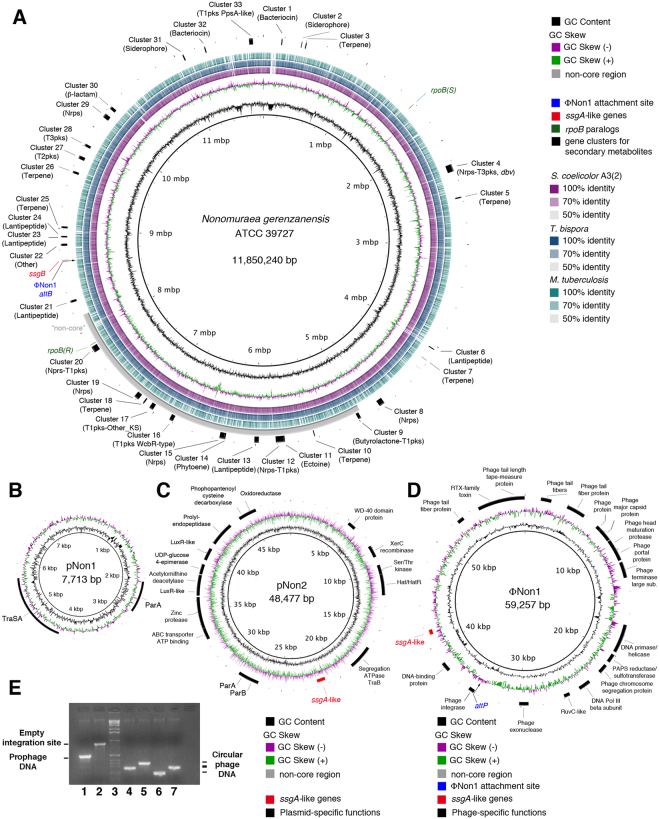
Figure 1.
Schematic representation of the circular chromosome of N. gerenzanensis, and maps and features of the extra-chromosomal elements. (A–D) Representations of the chromosome (A), plasmids pNon1 (B) and pNon2 (C), and the plasmid-phage element ΦNon1 (D) were obtained by the Blast Ring Image Generator (BRING). In panel A the N. gerenzanensis ATCC 39727 chromosome sequence was aligned with those of reference actinobacteria S. coelicolor A3(2), Mycobacterium tuberculosis H37Rv and T. bispora DSM 43833 as shown. In addition to GC content and GC skew, location of the rRNA operons, the gene clusters coding for secondary metabolites, the two rpoB(S) and rpoB(R) paralogs, ssgB (ssgA-like) gene, and the ΦNon1 attachment site are shown in panel A. The grey arc marks position of the “non-core” region. In panels B–D location of several phage/plasmid elements is shown including the ssgA-like genes in pNon2 and ΦNon1 (C,D). (E) Chromosomally integrated prophage ΦNon1 DNA (lane 1), empty chromosome attB integration site (lane 2) and circular ΦNon1 DNA (lanes 4–7) detected by PCR with specific primers.