Figure 1.
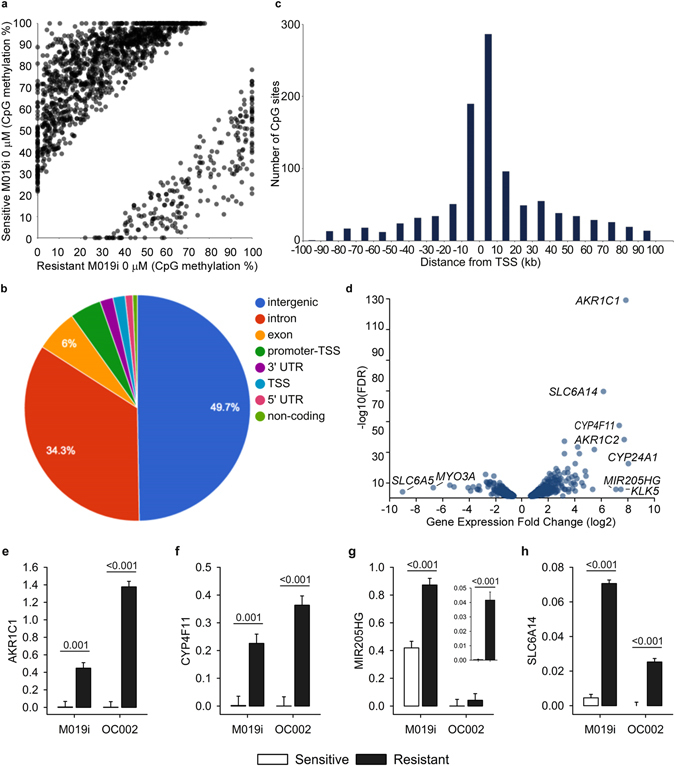
CpG methylome and transcriptome differences between cisplatin sensitive and resistant ovarian cancer lines. DNA methylomes of the cells sensitive or resistant to cisplatin were profiled with Reduced Representation Bisulfite Sequencing and transcriptomes with mRNA-sequencing. In (a) are the CpG sites with coverage ≥ 10 and minimum methylation difference of 20% (qval ≤ 0.05) in M019i cells, (see also Supplementary Table S1), (b) the distribution of differentially methylated cites in genomic regions, (c) distance of the differentially methylated sited from the closest transcription start sites, (d) the transcriptome differences (minimum absolute FC = 1.5, FDR ≤ 0.05) between cisplatin sensitive and resistant ovarian cancer cells (M019i), (e–h) qRT-PCR validation of AKR1C1, CYP4F11, CYP24A1, MIR205HG, and SLC6A14 differences in M019i and OC001 cells (y-axis: relative expression level).
