Figure 1.
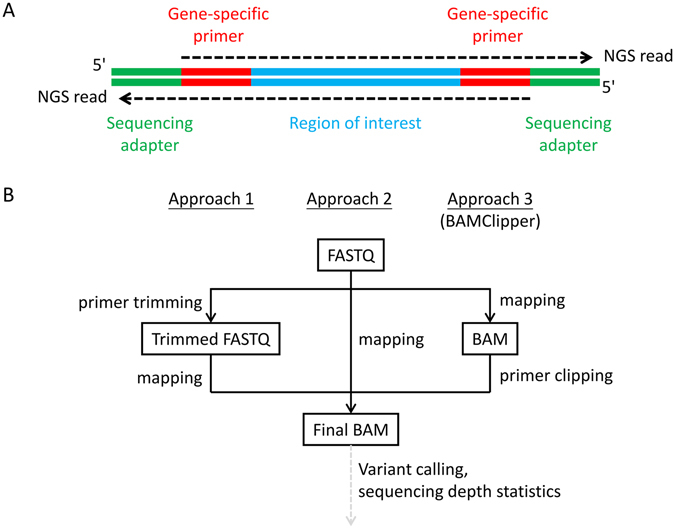
Amplicon library design and bioinformatics approaches of handling gene-specific primers. (A) Gene-specific primer sequences are present as part of NGS reads. The observed primer sequences are usually identical to reference genome sequence but may be slightly different due to errors in primer synthesis and/or sequencing. Common read mapping tools are not aware of the amplicon library design and thus map the primers as if they are part of region of interest that lies in between. Although sequencing adapters may exist as part of NGS reads (depending of amplicon length and sequencing read length), they will become soft-clipped after mapping due to the lack of similarity to reference genome by design. Soft-clipped part of alignments is usually ignored by downstream processing. (B) In primer handling approach 1, primer sequence was trimmed from sequencing reads (in FASTQ format) and the shorter trimmed reads are mapped to give BAM alignments for downstream variant calling and quality control such as sequencing depth statistics. In approach 2, original reads are directly mapped that primers are present in BAM alignments as if they are part of region of interest. In approach 3 represented by BAMClipper, reads are also directly mapped but BAM alignments are further processed to soft-clip primer sequences as if they were sequencing adapters.
