Figure 4.
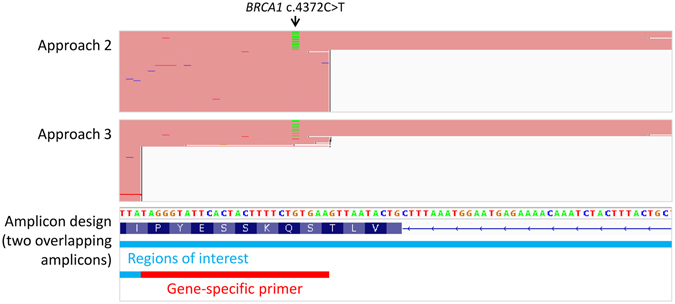
Dilution of variant allele frequency when primers are not clipped after mapping. NGS read alignments in BRCA1 c.4372C>T region from primer handling approaches 2 and 3 are shown in conjunction with the amplicon design and reference genome sequence. The c.4372C>T mutation is located in the region of interest of one amplicon and gene-specific primer site of another amplicon. Since primer sequences retained in approach 2 contributed to wild-type allele frequency, VAF of c.4372C>T was in turn underestimated by 82% (9% in approach 2 and 51% in approach 3).
