Figure 4.
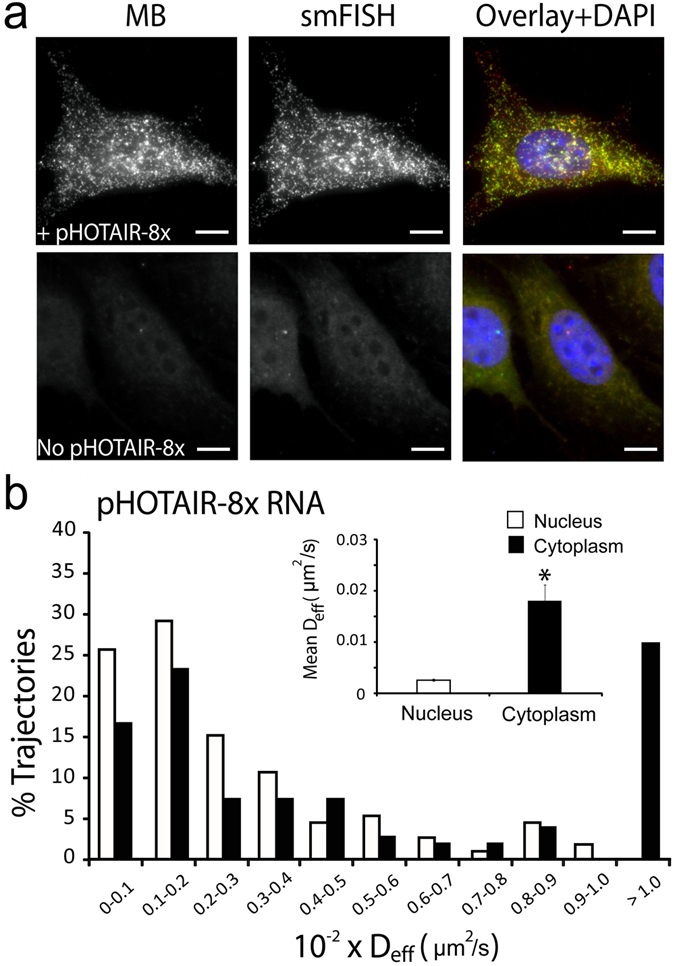
MB-based imaging of single HOTAIR transcripts in the nucleus and the cytoplasm. (a) Images of anti-repeat MBs for the detection of single pHOTAIR-8x transcripts. Following microporation of NIH3T3 cells with or without pHOTAIR-8x with 5 µM of anti-repeat MBs, the cells were fixed and permeabilized and smFISH was performed to assess the accuracy of MBs for detecting single RNA transcripts. Representative maximum intensity projection images of MB and smFISH signals are shown. Note that in NIH3T3 cells not transfected with pHOTAIR-8x, lack of smFISH signal shows the specificity of the smFISH probes for the human HOTAIR transcript used. (Scale bar, 10 µm) (b) Diffusion kinetics of single pHOTAIR-8x lncRNA transcripts in HeLa cells. HeLa-HOTAIR-8x cells were microporated with 5 µM anti-repeat MBs, and time-lapse images were acquired 8 h after microporation. Single particle tracking analysis was performed to determine the diffusion coefficient of individual RNAs as described in Materials and Methods. The distribution of diffusion coefficients of single mobile pHOTAIR-8x RNAs in the nucleus (n = 113 tracks) and in the cytoplasm (n = 255 tracks) analyzed from 30 cells. Inset shows the mean ± SE diffusion coefficients.
