Figure 1.
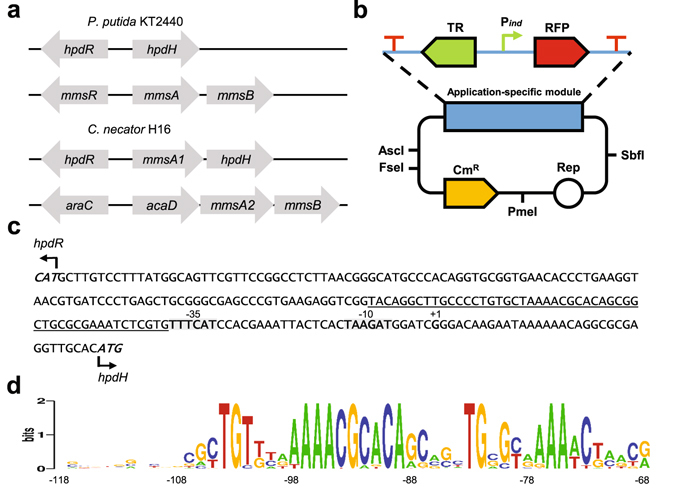
Identification of putative 3-HP-responsive genes in P. putida KT2440 and C. necator H16. (a) Operons putatively involved in 3-HP metabolism in P. putida KT2440 and C. necator H16. They are composed of the TR coding sequences hpdR, mmsR or araC, and divergently transcribed genes which are putatively required for 3-HP degradation: hpdH, 3-hydroxypropionate dehydrogenase; mmsA, methylmalonate-semialdehyde dehydrogenase; mmsB, 3-hydroxyisobutyrate dehydrogenase and acaD, acyl-CoA dehydrogenase. (b) Schematic illustration of the modular reporter system. It contains the four unique restriction sites AscI, FseI, PmeI and SbfI which are used for modular assembly. The application-specific module harbours the rfp reporter gene and the switchable system composed of transcriptional regulator and inducible promoter. (c) The P. putida KT2440 hpdR/hpdH intergenic region. Translational start sites are bold and italicised. The predicted hpdH −35 and −10 regions are bold and highlighted in grey. The nucleotide sequence between position −118 and −68 relative to the P. putida KT2440 hpdH translational start site is underlined. (d) A sequence similarity motif was generated which corresponds to the underlined sequence. It represents highly conserved nucleotides in the hpdR/hpdH intergenic regions of forty analysed Pseudomonas species.
