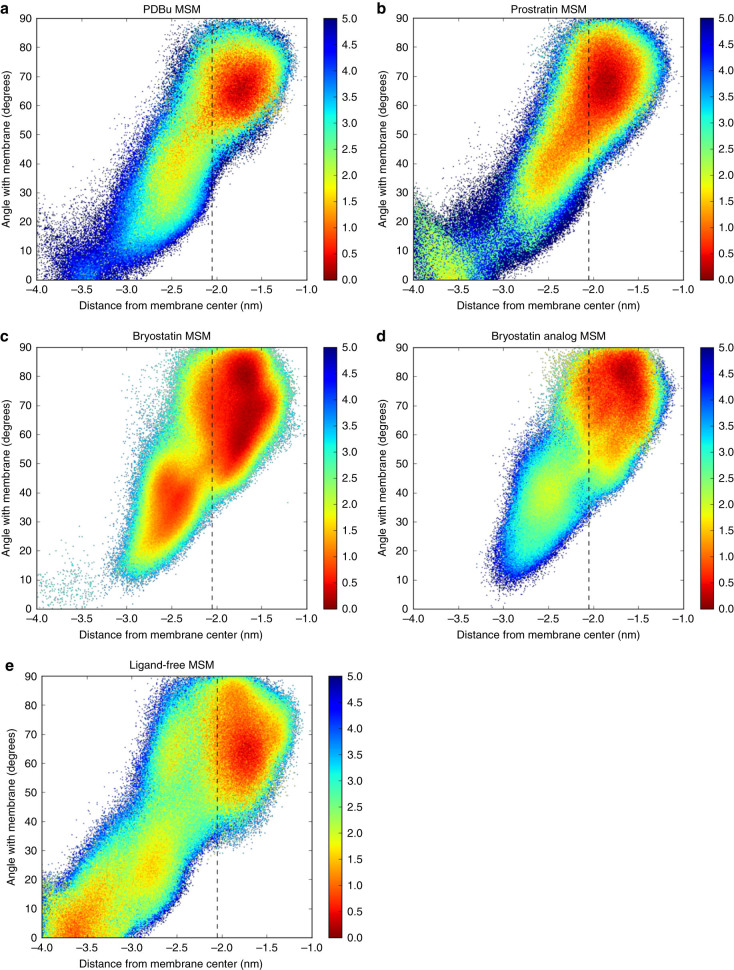
Fig. 3.
Free energy as a function of depth and angle of peptide in membrane. The panels represent the free energies of the peptide in the presence of various ligands (a–d) and without a ligand (e), reported in kcal mol−1. Note that bryostatin c and (to a lesser extent) prostratin b possess two low-free energy wells, one shallow and one deep, while PDBu a and bryolog 3 d only show one free energy minimum, deeply embedded in the membrane. The dashed line indicates the pseudo plane of phosphorus atoms in the bottom leaflet, and distance 0 is the plane through the center of the membrane. Distance is measured from an average of the positions of the alpha carbons of M9, T12, L21, and V25, all atoms near the binding site. The angle is found by first creating a line through this point and an average of the positions of the alpha carbons of F3, G35, N48, and the zinc ion coordinated by H1, C31, C34, and C50, and calculating the angle of this line with the membrane plane. This line goes approximately through the middle of the relatively cylindrical peptide. The range of the 95% confidence interval as determined by bootstrapping simulations was less than 0.1 kcal mol−1 for all configurations and all ligands. See Supplementary Fig. 4 and the Supplementary Discussion for further discussion of error bars for these free energies