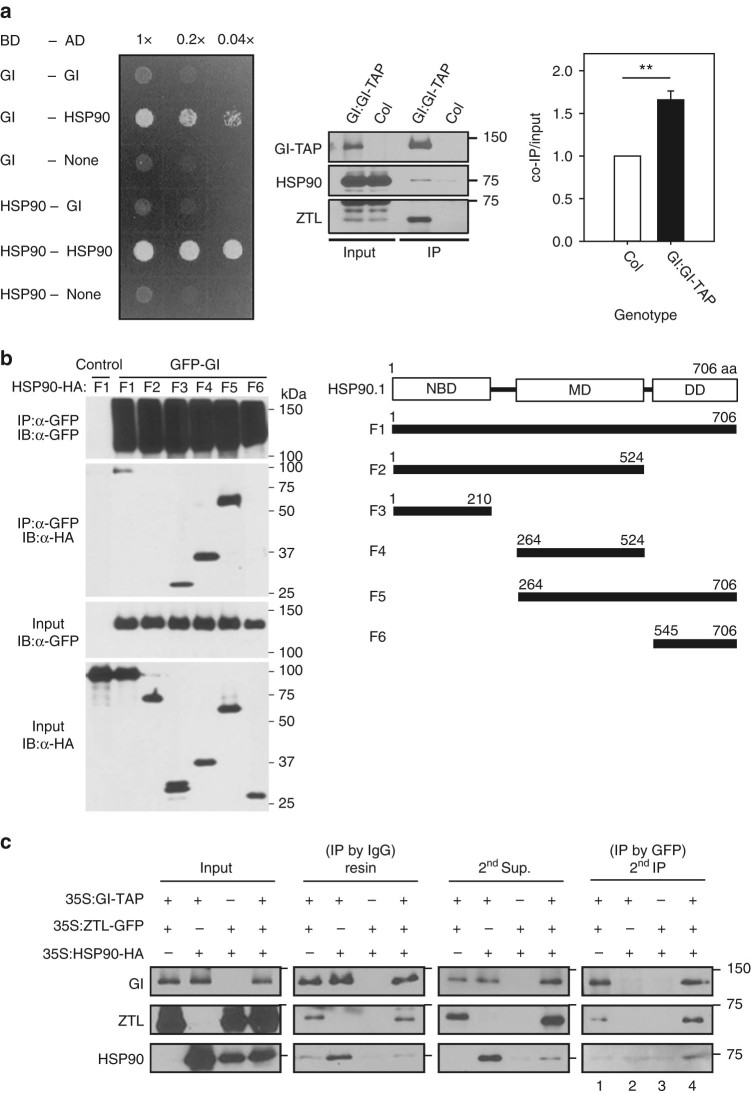
Fig. 4.
GI complex formation with ZTL and HSP90 in vivo. a GI and HSP90 interact directly in yeast (left panel) and in planta (right panel). Full-length GI and HSP90 protein interaction via yeast two-hybrid was determined by growth on leucine deficient media. Transgenic Arabidopsis expressing GI:GI-TAP immunoprecipitates with endogenous HSP90 and ZTL (right panels). Untransformed Col controlled for non-specific HSP90 interaction. Quantification of HSP90 in GI-TAP immunoprecipitations (IPs) (far right panel; mean ± s.e.m.; **P < 0.01) (n = 4). b HSP90 deletion interactions with GI. Agrobacteria harboring GI-GFP or HSP90 and its respective deletions tagged with 3 × HA were co-infiltrated into N. benthamiana leaves. Anti-GFP (IP) were followed by detection of co-immunoprecipitated HSP90-HA and respective deletions. Representative of three trials with similar results. Right panel: HSP90 domain structure and respective deletion scheme. IP immunoprecipitating antibody, IB immunoblot antibody, NBD nucleotide-binding domain, MD middle domain, DD dimerization domain. c GI, ZTL, and HSP90 form a tripartite protein complex in planta. Sequential co-immunopreciptations used GI-TAP in the primary IP followed by IP of ZTL-GFP (anti-GFP ab) from the protease-released supernatant (2nd Sup.). The final detection of HSP90-HA (anti HA ab) in lane 4 indicates HSP90-HA associated with ZTL-GFP from the first IP. N. benthamiana leaves were triply co-infiltrated with 35S:GI-TAP/35S:HSP90-HA/35S:ZTL-GFP simultaneously or in all pairwise combinations. Representative of three independent trials