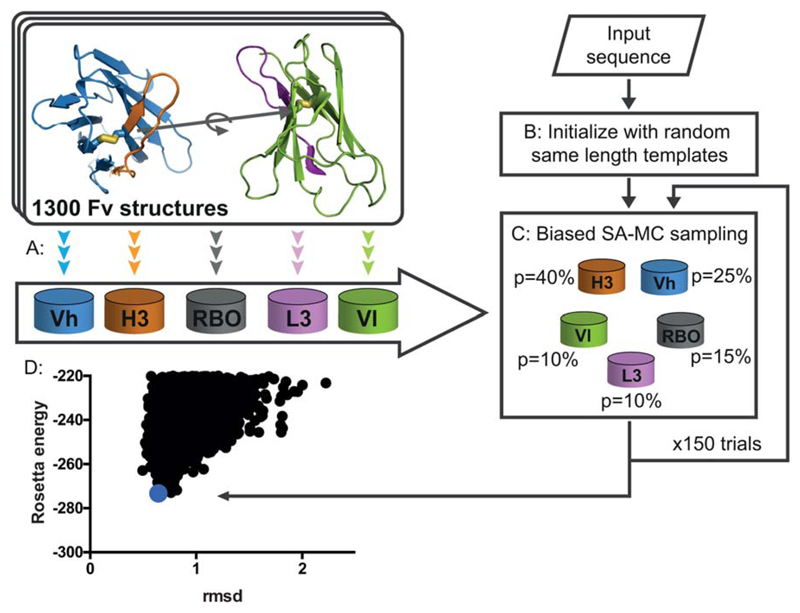
Figure 1.
AbPredict: A method for combinatorial backbone modeling of antibodies. (A) Database preparation: The antibody variable domain (Fv) is segmented into regions comprising CDRs L3 and H3 (magenta and orange, respectively), and regions comprising much of the V segments, Vh (blue) and Vl (green), including CDRs 1 and 2.9 In a pre-computation step the backbone torsion angles of each segment as well as the RBO between light and heavy chains (gray) are stored in five databases for efficient lookup during modeling simulations. (B & C) Schematic description of a structure prediction trajectory: starting from the input sequence an arbitrary combination of backbone and rigid-body degrees of freedom is chosen from the databases and 150 simulated-annealing Monte Carlo (SA-MC) steps are conducted to identify low-energy conformations. (D) 3,000 independent trajectories are run to produce an energy landscape, where every point is the lowest-energy structure from an individual modeling trajectory and carbonyl root mean square deviation (rmsd) is measured to the experimentally determined structure.