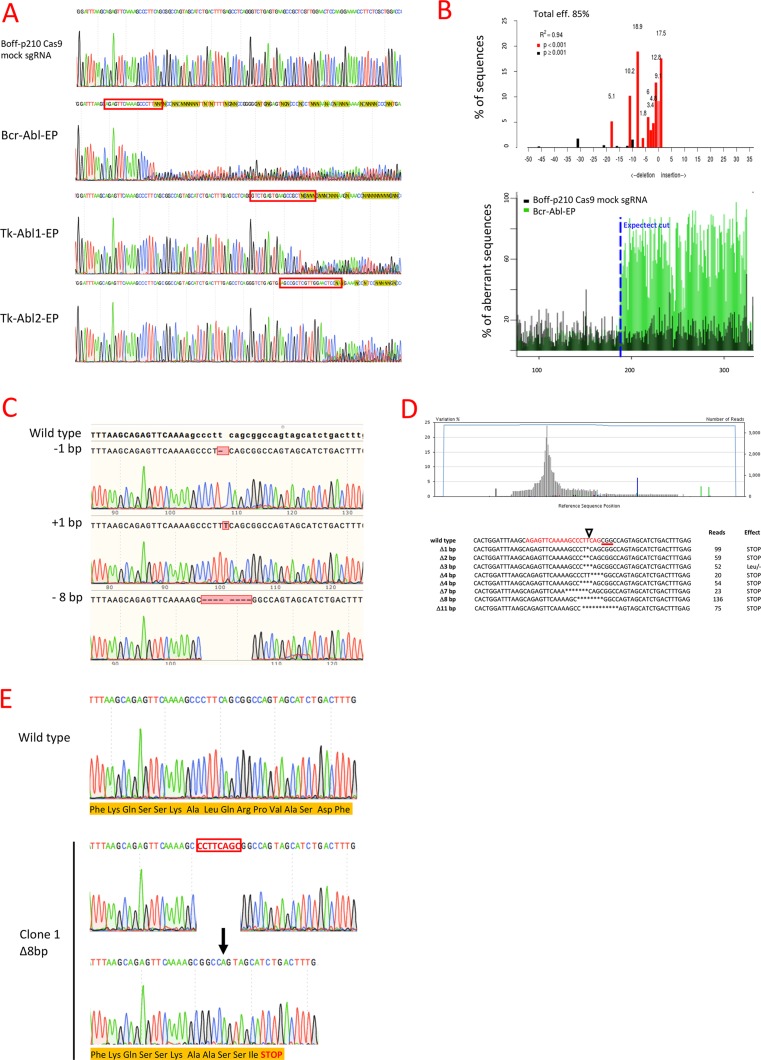
Figure 2. Genome editing of BCR/ABL in the Boff-P210 cell line.
(A) Sanger sequencing of the BCR/ABL fusion region in Boff-p210 cells. The Boff-p210 cells expressing mock sgRNA, used as a control, had a wild type sequence, while cells expressing Bcr-Abl sgRNA (Bcr-Abl-EP), Tk-Abl1 sgRNA (Tk-Abl1-EP) and Tk-Abl2 sgRNA (Tk-Abl1-EP) showed a mixture of sequences around the expected Cas9 cleavage point. (B) TIDE decomposition algorithm analysis of the edited sequence in Bcr-Abl-EP cells, showing high editing efficiency at the expected cleavage point. The lower panel illustrates the aberrant sequence signal in Boff-p210 cells (black) and Boff-p210-edited cells (green) and the expected cleavage site (vertical dotted line). (C) DNA sequences of DNA post-PCR cloning colonies of Boff-p210-edited cell DNA. We compare three induced mutations with the original sequence. (D) Next-generation sequencing of the transgene target region. The upper panel shows the percentages of each base pair in the sequenced region. The lower panel shows commonly sequenced variants for CRISPR-targeted Bcr/Abl transgene in Boff-p210 cells. The sgRNA-target site is displayed in red, the PAM sequence as text underlined in red, and the cleavage site is indicated by a triangle. The number of reads per variant and the predicted effect of the mutation are shown. (E) In silico analysis of single cell-derived clone. Clone 1 showed a 8 bp deletion resulting in a stop codon and in a premature end of translation. It was selected to establish the Bcr-Abl-SC cell line.