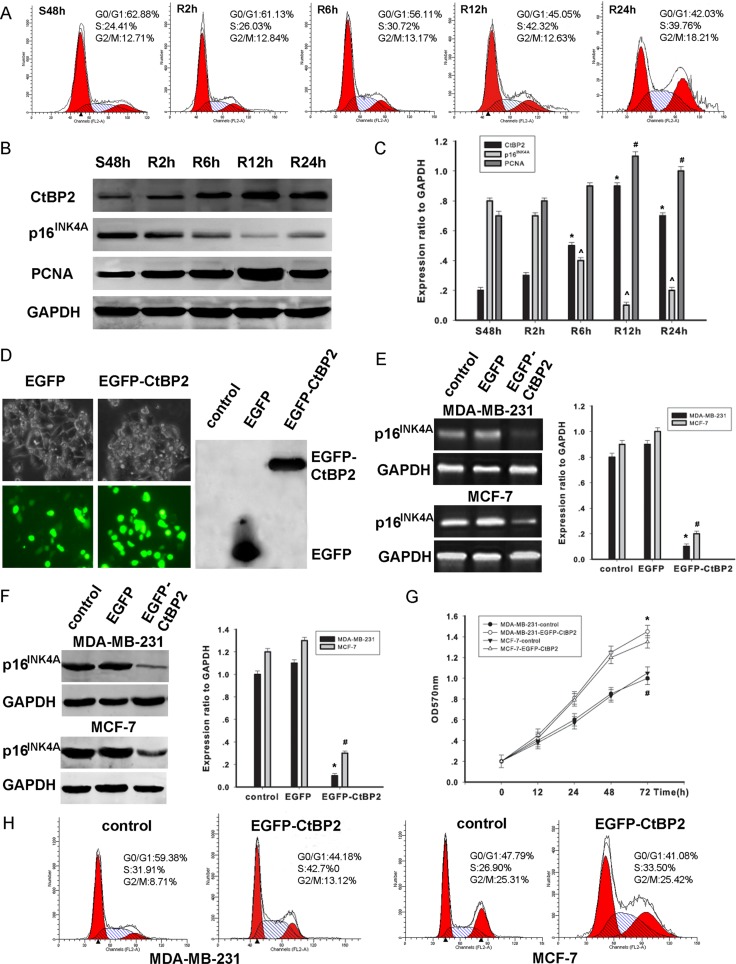
Figure 4. CtBP2 plays a proliferative role in breast cancer cells.
(A) Flow cytometry analysis of cell cycle progression in MDA-MB-231 cells. Cells that were synchronized at G1 progressed into the cell cycle 0, 2, 6, 12, and 24 h after serum stimulation. Finally, most of the cells entered S phase. (B) MDA-MB-231 cells were serum starved for 48 h (S48h). Upon serum stimulation, cell lysates were prepared and analyzed by Western blot using antibodies against CtBP2, p16INK4A, PCNA and GAPDH. GAPDH was used as a control for loading and protein integrity. (C) The bar graph demonstrates the ratio of CtBP2, p16INK4A and PCNA proteins to GAPDH at each time point, as determined by densitometry. The data are represented as the mean ± SEM (n = 3, *^#P < 0.01, compared with control: S48 h). S: serum starvation; R: serum stimulation. (D) Light microscopy shows that pcDNA3.1-EGFP and pcDNA3.1-EGFP-CtBP2 are expressed in MDA-MB-231 cells. Proteins were analyzed by Western blot using an anti-EGFP antibody. (E) MDA-MB-231 and MCF-7 cells were transfected with pcDNA3.1-EGFP- CtBP2 or nothing (control). RT-PCR shows transcriptional levels of the p16INK4A gene 48 h post-fection, and GAPDH was used as a loading control. The data are means ± SEM *#P < 0.01, compared with the control group. (F) MDA-MB-231 and MCF-7 cells were transfected with the pcDNA3.1-EGFP-CtBP2, or control, as indicated. Protein expression of p16INK4A and GAPDH was analyzed by Western blot. Data are presented as means±SEM *#P < 0.01, compared with the control group. (G) MDA-MB-231 and MCF-7 cells were transfected with either pcDNA3.1-EGFP-CtBP2 or control. Cell growth of the transfected cells was assessed by the CCK-8 cell viability assay. The data are presented as the mean±standard error of three experiments. (H) Cell cycle analysis was performed by staining CtBP2 overexpressing MDA-MB-231 and MCF-7 cells with PI.