Figure 2.
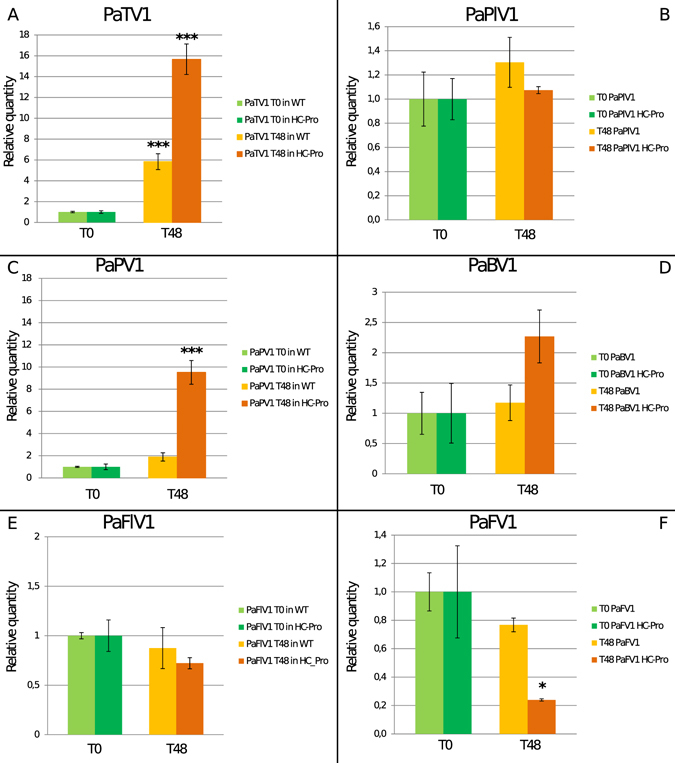
Comparison of virus accumulation in protoplasts from wild type (WT) and HC-Pro Nicotiana benthamiana. Quantitative Reverse transcriptase PCR (qRT-PCR) data from WT and HC-Pro N. benthamiana protoplasts were used to calculate fold change of viral RNAs at T0 and T48 time points. In panels A data for the Totivirus Penicillium aurantiogriseum totivirus 1 (PaTV1) are reported: for WT protoplasts we observed ca. 5 times more RNA at T48 (yellow bar) time point and ca. 15 times more RNA in HC-Pro protoplasts (orange bar). In panels C quantification of the Partitivirus Penicillium aurantiogriseum partitivirus 1 (PaPV1): no statistically significant differences were observed for WT protoplasts, but ca. 10 times more viral RNA was observed in case of HC-Pro protoplasts. Panels B, D, E and F show data for the other viruses: none of them displays a statistically significant increase in virus RNA load. *P < 0.05 **P < 0.01 and ***P < 0.001.
