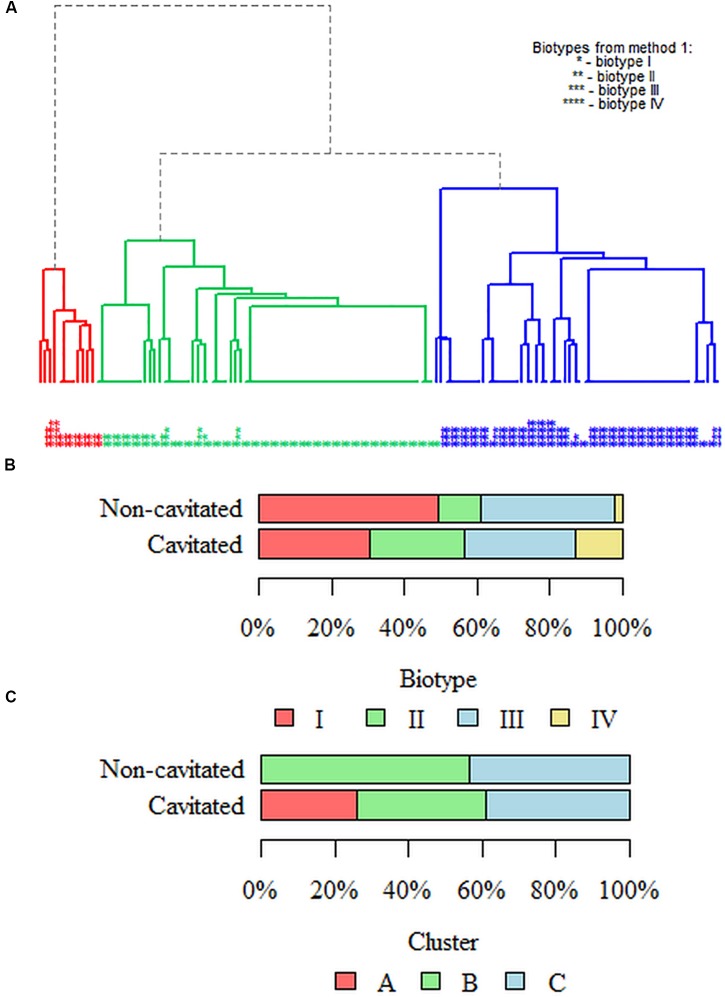
FIGURE 1.
Dendrogram with three marked biotypes (clusters) of the studied Streptococcus mutans population. The criterion for enzyme biotyping in the arbitrary method was the selection of chosen enzymes based on the analysis of their activity in the population of the tested strains. We have included those enzymes whose activity in the test population is in the 15–85% quartile deviation. Hence, there are three enzymes: inulin, melibiose and tagatose, giving eight profiles eventually resulting in four biotypes (I–IV) marked on the chart with stars (∗–∗∗∗∗). Three clusters [the main branches in the figure colored red (cluster A), green (cluster B) and blue (cluster C)] were obtained by clustering using the hierarchical method. In the unsupervised clusterization method (without a priori knowledge), data were divided into clusters so that each group was as homogeneous as possible (the strains within the groups are similar), and the clusters were mutually different (strains from different groups have as few common features as possible). For the best-fitted dendrogram, strains were differentiated on the basis of inulin, melibiose and tagatose activity. This allowed for the differentiation of three clusters – the main branches in the figure in different colors and described in the text as A–C. The distribution of enzymatic profiles obtained by the arbitrary method (method no. 1) is also shown. The red cluster (A) is basically biotype II; green is biotype I, and blue is biotype III.