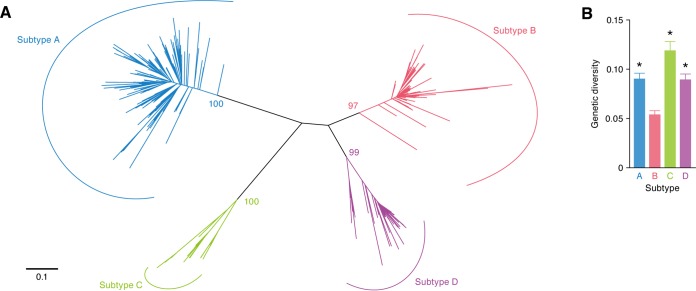
FIG 1.
Less diversity of FIV subtype B. The sequences of the V3-V5 region of FIV env (subtype A, n = 153; subtype B, n = 100; subtype C, n = 24; subtype D, n = 49) were extracted from the GenBank/EMBL/DDBJ sequence database (the accession numbers used are available upon request). (A) Phylogenetic tree of FIV env. This phylogenetic tree was constructed using the ML method and displays an evolutionary relationship among the FIV sequences used in this study. Each line, called a “branch” of the tree, represents one FIV sequence. The longer branch lengths correlate with larger accumulations of mutations in the sequences. The scale bar indicates 0.1 nucleotide substitution per site. The bootstrap values are indicated on each node. (B) Viral genetic diversity. These values were calculated as described in Materials and Methods. Statistical analyses were performed using Bonferroni's multiple-comparison test. *, P < 0.01 versus subtype B.