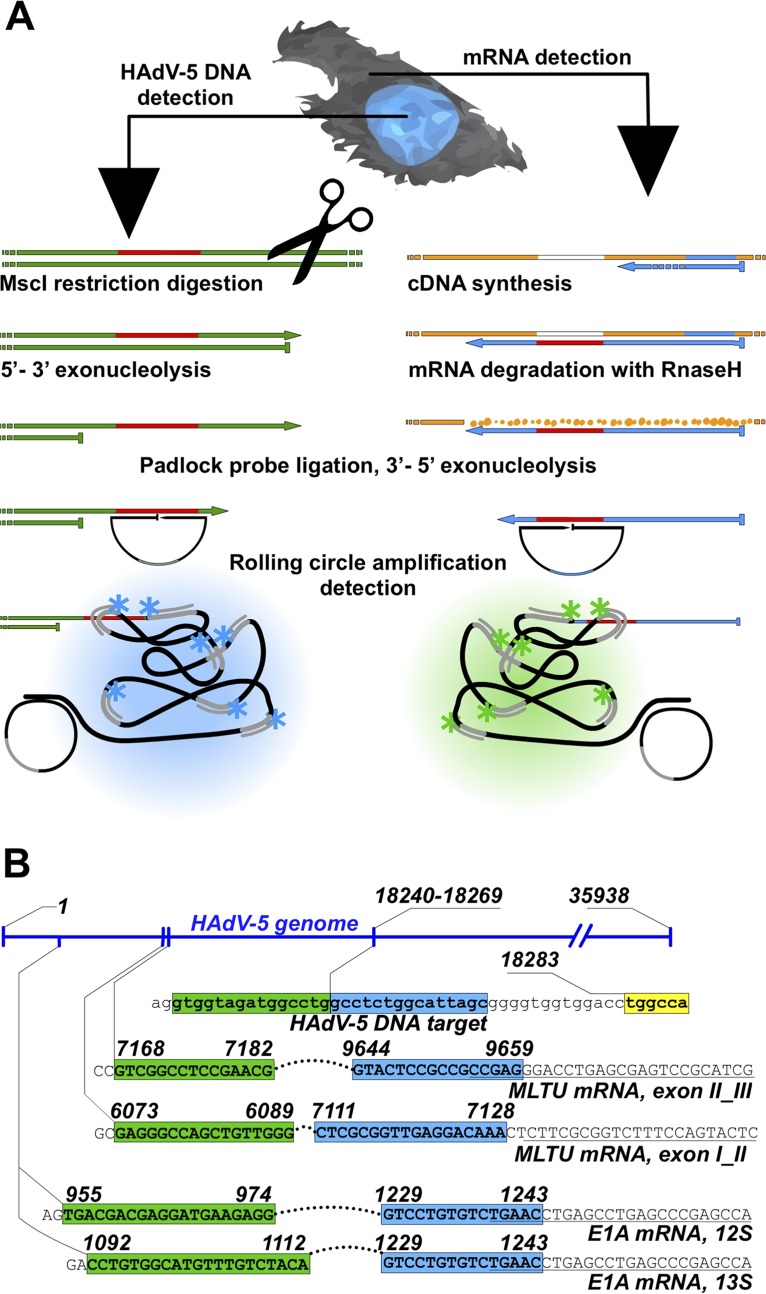
FIG 1.
HAdV-5 DNA and mRNA detection by rolling-circle amplification (RCA). (A) Simplified schematic overview of concurrent viral DNA and mRNA detection. Viral mRNA is converted to cDNA by reverse transcription, followed by mRNA degradation by RNase H. HAdV-5 DNA endonucleolytic cleavage with MscI and λ-exonuclease digestion expose ssDNA target sequence for padlock probe (PLP) binding. Target annealed PLPs are circularized with DNA ligase and amplified by RCA. The generated rolling-circle product (RCP) is visualized upon binding of fluorophore-conjugated oligonucleotides under a fluorescence microscope. Arrows represent 3′ extremities, and flat ends represent 5′ extremities. (B) PLP target sites on HAdV-5 DNA and mRNAs. The E1A mRNA and MLTU mRNA PLP target sequences (uppercase letters) are presented on the corresponding HAdV-5 genome regions. 5′ and 3′ arms of the PLP target sequences are indicated in green and blue, respectively. The MscI restriction site adjacent to the HAdV-5 genomic DNA padlock probe sequence target is in yellow. RT primer binding sites adjacent to E1A and MLTU target sites (italic). HAdV-5 genome numbering is based on NCBI reference sequence AC_000008.1.