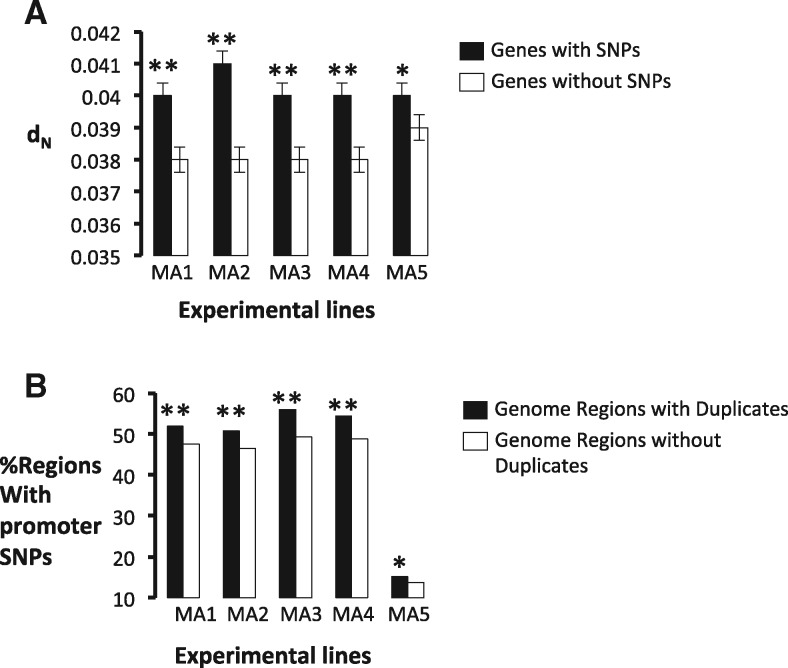
Fig. 2.
—Experimental evolution of S. cerevisiae reveals that mutational genome hotspots are traps for duplicates. We identified mutations in the coding region and 600-nucleotide genome regions upstream genes in S. cerevisiae and calculated the nonsynonymous nucleotide substitutions (dN) between S. cerevisiae and its close phylogenetic relative S. paradoxus for the six genes surrounding that gene with a SNP in its promoter. (a) Genes that have fixed a SNP in the promoter regions (black columns) in each of the five experimentally evolving line (MA1–MA5) exhibit significantly higher (* indicates P < 0.01; ** indicates P < 0.001) dN than those without SNPs (white boxes). (b) Genome regions that contain duplicates (GRDs) exhibit more SNPs than in the promoter (black columns) fixed during the evolution experiment of each line (MA1 to MA5) (* indicates P < 0.01; ** indicates P < 0.001) than genome regions that do not contain duplicates (GRSs) (white columns).