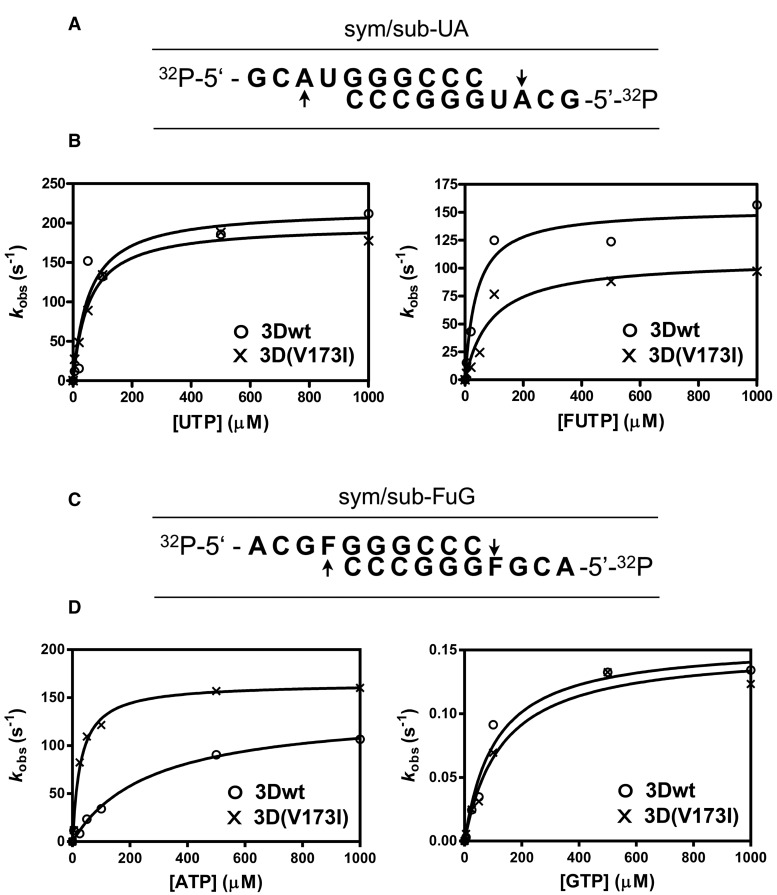
Fig. 2.—
Presteady-state kinetics of nucleotide incorporation into sym/sub-UA and sym/sub-FuG by 3Dwt and 3D(V173I). (A) Sequence of 5′-end-labeled, annealed sym/sub-UA. Arrows indicate the template residue at which nucleotide incorporation is measured. (B) 3D (0.5 µM active sites) was preincubated at 37 °C with sym/sub-UA (0.5 µM duplex) and ATP (10 µM) for 900 s to allow the formation of 3D-RNA product complex, with AMP incorporated at the first position. The 3D-RNA product was then mixed with the indicated concentration of UTP or FUTP using a rapid chemical quench-flow apparatus, and reactions were quenched by the addition of EDTA (0.3 M). Independent time points at 0, 0.01, 0.05, 0.1, 0.25, 0.5, 1, and 2 s were taken for each nucleotide concentration tested. Time courses at fixed nucleotide concentrations were fit to an exponential curve to obtain the observed rate constant for nucleotide incorporation at the second position, kobs. The observed rate constants were then plotted as a function of nucleotide concentration, and the data were fit to a hyperbola to obtain kpol and Kd,app. Note that the scale at ordinate is different in the two panels. (C) Sequence of 5′-end-labeled, annealed sym/sub-FuG (Fu means FU). Arrows indicate the template residue at which nucleotide incorporation is measured (FU is written F). (D) Incorporation of AMP or GMP; no nucleotide was preincubated with 3D and RNA. Procedures are those described in (B). For the assays with GTP, the concentration of 3D used was 1 µM active sites, and no rapid chemical quench-flow apparatus was needed. Duplicate samples at 0, 10, 30, 60, 120, 300 and 900 s time points were taken for each nucleotide concentration tested. Note that the scale in ordenate is different in the two panels. Procedures are further detailed in Materials and Methods.