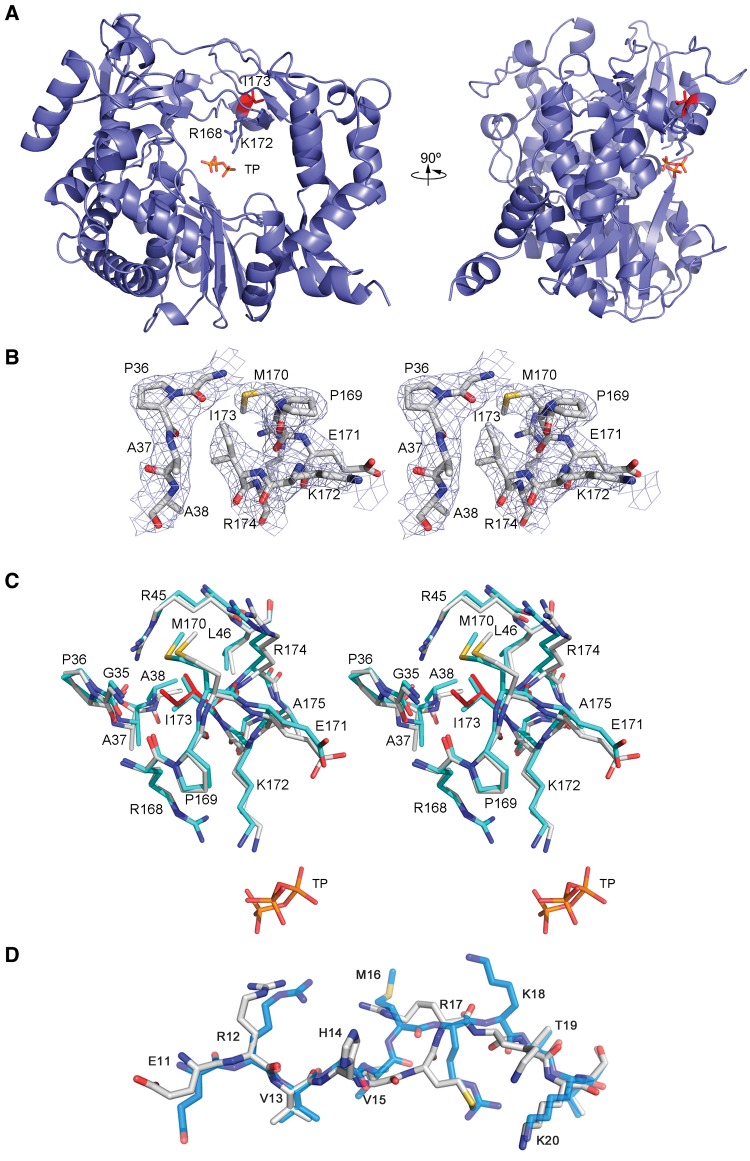
Fig. 3.—
Structure of FMDV-3D(V173I). (A) The left and right panels show two views of 3D(V173I) protein rotated by 90°. The polymerase is depicted in blue ribbons with substituted amino acid I173 shown in sticks in red. The triphosphate moiety of the bound ATP and the motif F contacting side chains R168 and K172 are also shown as sticks and explicitly labeled. (B) Stereoview of σA-weighted |Fo| − |Fc| electron density map (contoured at 3σ) around the mutated residue I173. The substituted residues and surrounding amino acids were omitted from the phasing model. The model is placed inside in ball and stick representation and colored in atom type code. (C) Stereoview of the structural changes around the mutated I173 residue. 3D(V173I) polymerase is shown in white (with I173 depicted in red) and the superimposed 3Dwt in cyan (TP refers to the ATP triphosphate moiety). (D) Superimposition of the N-terminal region (residues E11–K20) of the FMDV-wt polymerase (cyan) and the FMDV-3D(V173I) polymerase (white). Information on data collection and refinement statistics is given in supplementary table S3, Supplementary Material online. The PDB accession codes for the 3Ds represented here are 1WNE for 3Dwt and 5DTN for 3D(V173I).