Figure 2.
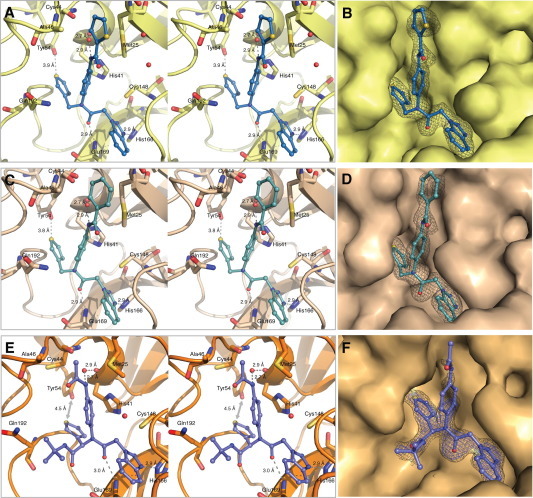
X-ray crystal structures of 1A, 2A and 3B in complex with HKU4-CoV 3CLpro. Inhibitors are colored according to atom type and shown in ball and stick representation. Wall-eye stereo-view is shown in panels A, C, and E. Water molecules are shown as red spheres. Hydrogen bonds are shown as gray dashes with distances between heteroatoms labeled (Å). Interatomic distances (Å) are displayed using a gray arrow. Electron density omit maps (Fo − Fc) are shown in grey mesh and are contoured to +3.0σ around the inhibitor only. The binding orientation of each inhibitor is the same in the active site of each monomer of the dimer in the asymmetric unit of HKU4-CoV 3CLpro, therefore only one active site is shown for clarity. (A) HKU4-CoV 3CLpro (pale yellow) in complex with 1A (sky blue), PDB 4YOI, (B) 1A electron density omit map, (C) HKU4-CoV 3CLpro (wheat) in complex with 2A (pale cyan), PDB 4YOJ, (D) 2A electron density omit map, (E) HKU4-CoV 3CLpro (light orange) in complex with 3B (slate), PDB 4YOG, (F) 3B electron density omit map contoured at +3.0σ (blue) and +2.0σ (grey).
