Abstract
Background
A large proportion of the Ontario population lives with a diagnosed mental illness. Nearly 5% of Ontarians have major depressive disorder, and another 5% have another type of depressive disorder, bipolar disorder, schizophrenia, anxiety, or some other disorder not otherwise specified. Medications are commonly used to treat mental illness, but choosing the right medication for each patient is challenging, and more than 40% of patients discontinue their medication within 90 days because of adverse effects or lack of response. The Assurex GeneSight Psychotropic test is a pharmacogenomic panel that provides clinicians with a report to guide medication selection that is unique to each patient based on their individual genetic profile. However, it is uncertain whether guided treatment using GeneSight is effective compared with unguided treatment (usual care).
Methods
We performed a systematic review to identify English-language studies published before February 22, 2016, that compared GeneSight-guided care and usual care among people with mood disorders, anxiety, or schizophrenia. Primary outcomes of interest were prevention of suicide, remission of depression symptoms, response to depression therapy, depression score, and quality of life. Secondary outcomes of interest were impact on therapeutic decisions and patient and clinician satisfaction. Risk of bias was evaluated, and the quality of the evidence was assessed using the Grading of Recommendations Assessment, Development, and Evaluation (GRADE) working group criteria.
Results
Four studies met the inclusion criteria. These studies used a version of GeneSight that included the CYP2D6, CYP2C19, CYP1A2, SLC6A4, and HTR2A genes; one of the studies also included CYP2C9. Patients who received the GeneSight test to guide psychotropic medication selection had improved response to depression treatment, greater improvements in measures of depression, and greater patient and clinician satisfaction compared with patients who received treatment as usual. We observed no differences in rates of complete remission from depression. The findings were based on GRADE assessment of low to very low quality evidence, and the body of evidence had several limitations: the included studies used an older version of GeneSight and were limited to a population with major depression, so results may not be generalizable to other versions of the test or different populations such as patients with anxiety or schizophrenia.
Conclusions
There is uncertainty about the use of GeneSight Psychotropic pharmacogenomic genetic panel to guide medication selection. It was associated with improvements in some patient outcomes, but not others. As well, our confidence in these findings is low because of limitations in the body of evidence.
BACKGROUND
Health Condition
Mental illness, specifically mood disorders, anxiety, and schizophrenia, can affect daily living and may be accompanied by fatigue, insomnia, sudden weight loss, and an overall depressed mood, among other symptoms.1 Approximately 10% of Ontarians live with mental illness, and 4.8% live with major depression.2 Of people who contact the emergency department about a mental illness, a third (34%) do so because of anxiety, and another 18% because of other mental illnesses.2 Of people admitted to hospital because of mental illness, 12.4% have bipolar disorder and 28.4% have major depressive disorder.2 People with one form of mental illness often experience other forms as well. For example, as many as 70% of people who have generalized anxiety disorder also have another mental illness, and 45% of people with anxiety also live with depression at some point in their lifetime.3
There are some differences in equity related to rates of mental illnesses and people's experience of them. For example, major depression affects 5.8% of women and only 3.8% of men.2 Bipolar disorder is more commonly seen among younger people (2.4% of people aged 15 to 24 years vs. 1.7% of people older than 45 years).2 As well, people from low-income areas are at greater risk of developing a mental illness than people from high-income areas.2
Variation in access to care is another equity consideration. At present, Toronto Central Local Health Integration Network (LHIN) has 63 full-time psychiatrists per 100,000 people, about three times more than Champlain, the next most concentrated LHIN, which has 24 psychiatrists per 100,000, and in great contrast to the Central West LHIN, which has only 4.7 psychiatrists per 100,000 people.2 As well, laboratory and genetic testing is not standardized across Ontario; people in one region may have access to different tests than people in another.
Treating mental illness is challenging, because people who have mental illness often avoid asking for professional help due to the stigma associated with a mental disorder. When they do seek treatment, they often need a combination of therapies, including psychotherapy (such as cognitive behavioural therapy), one or more medications (such as antidepressants), or both.4 Antidepressants are among the most commonly prescribed medications in young Canadians, used by 3.6% of males and 13% of females aged 6 to 24 years.5 They are also the most commonly prescribed medication for Canadian women, used by 13.7% of women aged 25 to 79 years.5 Antidepressant use is less common among Canadian men, but is still within the top five prescriptions, at 8.2% of those aged 44 to 64 years.5
The Canadian Network for Mood and Anxiety Treatments Clinical guidelines lists 17 different antidepressants as potential first-line medications for major depressive disorder, grouped by mechanism of action, including tricyclic antidepressants and selective serotonin reuptake inhibitors (SSRIs).6 However, choosing the right medication is a challenge. For example, even though SSRIs are listed as first-line treatments, they are effective in only a third of the patients treated with them; patients may need four or more different medications, plus dosage augmentations, to manage the symptoms of major depressive disorder.7
Of those who are treated, many do not respond sufficiently to treatment or experience a return of their symptoms.8 More than 40% of patients discontinue their medications within the first 90 days of therapy because of a lack of response, side effects, or both.6 Some of the more common side effects include drowsiness, headache, dry mouth, nausea, anxiety, and sexual dysfunction.6 There is also a risk of uncommon side effects such as seizures, bleeding in the upper gastrointestinal tract, and osteoporosis and fractures in the elderly.6
Clinical Need and Target Population
Individual variations in people's genetic code may cause them to respond differently to medications. They may experience differences in metabolism (how the drug is broken down and used in the body), differences in mechanism of action (how the drug works), and individual adverse effects. For example, a person classified as a poor metabolizer because of differences in their genetic coding may have a lower enzyme response rate and metabolize a medication more slowly.9,10 This would lead to higher concentrations of the medication in their system than a typical metabolizer would have, and thus, potential adverse effects. Ultra (or fast) metabolizers are less likely to experience side effects that lead to discontinuing the medication, but they may need a higher dosage to achieve a noticeable benefit.6
Genome-wide association studies have attributed as much as 42% of the variation in response to antidepressants to individual genetic differences, and other studies have demonstrated correlations with ethnicity and ancestral variations.10–12 It is believed that prescribing clinicians who know a patient's genetic predisposition could better target therapies, reduce the risk of adverse effects, and minimize the use of the health care system, services, and costs.13
More and more medications include pharmacogenomic biomarker information in their product labelling.14 The most common information related to psychotropic medications is a warning about dosage for poor metabolizers known to be associated with the CYP2D6 and CYP2C19 enzymes; for example, patients classified as poor metabolizers for CYP2C19 should receive a starting dose of citalopram at 60% of the dose for a typical metabolizer.14
As research in this area continues to evolve, the evidence is improving around some single genes to predict how effective a drug will be for an individual. According to one review, the best evidence for specific genes that can predict response to antidepressant efficacy are CYP2D6, CYP2C19, SLC6A4, HTR2A, BDNF, GNB3, FKBP5, and ABCB1.9 However, another review concluded that while there was some good evidence for certain individual candidate genes, there was no major effect of any single gene variant on antidepressant efficacy.15
Evaluating the usefulness of a combination of genes in a panel may be a more effective way to identify individual variations than assessing single genes alone16; this is believed to be due to the complex metabolic pathways that cause interactions between enzymes.16 Further adding to the complexity is growing evidence of the effects of environment on gene expression, biological variations in depression based on ancestry, and changes in enzyme metabolism with age.11,12,17–19 Using genetic testing to support health care decision-making is still a relatively new idea; it is still uncertain whether such tests can affect important patient outcomes.
Technology
GeneSight Psychotropic is a multi-gene, multivariant genetic test that combines genotype (a person's genetic profile), phenotype (a person's physical characteristics), and drug metabolism information in an algorithm to categorize included medications for each patient using a system of green, yellow, and red bins. “Green” medications are supported for use as usual, “yellow” for use with caution, and “red” for use with increased caution and more frequent monitoring by the prescribing clinician.20
The test can be ordered by any prescribing clinician, and the results of the test integrate a person's genetic information with each medication into an easy-to-read report that is proprietary to Assurex Health.20 The test is noninvasive and easy to administer: it requires only a cheek swab to collect a sample of a person's DNA. Results are provided within 36 hours, but rush service is available if needed.20
At present, the test is sent via courier to the Assurex Health laboratory. Samples are analyzed using primers developed by Assurex and existing DNA testing technology: the Luminex xTAG Multiplex Technology (Luminex, Austin, TX) and the Lonza FlashGel system (Lonza, Basel, Switzerland).20 The test has 100% sensitivity and specificity at concentrations of 20 ng/μL for all genes except SLC6A4, for which it is valid at 10 ng/μL.21 The test is also valid at concentrations of 5 ng/μL to determine CYP2D6 copy number variation.21
Eight genes were included in the version of the GeneSight Psychotropic test requested for review (Appendix 1). Of these, six were pharmacokinetic—members of the cytochrome P450 family of liver enzymes, which play an important role in overall drug metabolism. The other two were pharmacodynamic—associated with the serotonergic transporter and receptor genes, where variations can affect the mechanism of action of SSRIs. Although the requested version of GeneSight includes eight genes, the Assurex website currently lists a version that includes 12.20
Potential Value of GeneSight Psychotropic Testing
In a retrospective study conducted in 79 psychiatric patients with depressive disorder or anxiety, patients were classified using the GeneSight test and their health services utilization was analyzed, looking back 1 year into administrative data.13 Patients who took red-bin medications had more total health care visits and disability claims than patients who took yellow- or green-bin medications (Table 1). Patients who took red-bin medications also had significantly more medical absence days than patients who took green-bin medications alone (P = .043).13 The authors found significant correlations between the number of weeks a patient spent on a red-bin medication and higher numbers of health care visits (P = .05), as well as the total number of drugs taken and medical or other health care visits (P < .001).13 Taken together, these findings may demonstrate the potential for the GeneSight test to predict which treatment medication would be optimal for a person and support the avoidance of red-bin medications to minimize health care utilization, improve the benefit from selected therapies, and enhance patients' overall quality of life.13
Table 1:
Health Care Utilization by GeneSight Patient Classification
| Outcome Measure | Results by Patient Classification | Significance (Red-Bin Patients vs. Green- and Yellow-Bin Patients) | ||
|---|---|---|---|---|
| Red Bin (n = 9) | Yellow Bin (n = 28) | Green Bin (n = 39) | ||
| Total health care visits | 21.9 | 12.3 | 13.7 | P = .014 |
| Nonpsychiatric medical visits | 12.8 | 7.1 | 8.4 | P = .039 |
| Outpatient psychiatric visits | 8.9 | 5 | 5.1 | P = .145a |
| Number of disability claims | 0.56 | 0.11 | 0.15 | P = .013 |
| Medical absence days | 20.8 | 8.4 | 4.6 | P = .126 |
| Hospitalizations (days admitted) | NR | NR | NR | NSb |
| Emergency department visits | NR | NR | NR | NSb |
Abbreviations: NR, not reported; NS, not significant.
Calculated based on F statistic provided in the publication.
Reported as not statistically significant using a threshold of P > .05
Source: Winner et al.13
Another study compared the relative effectiveness of combinatorial genomics (evaluating the effects of several genes together) with single-gene phenotyping (evaluating the effects of one gene at a time). Altar et al16 used a cohort of patients who received usual care but also underwent genotyping with GeneSight. Patients were categorized as ultra-rapid, extensive, intermediate, or poor metabolizers based on single-gene phenotyping. Within those groups, patients were further categorized using combinatorial genomics and the GeneSight Psychotropic red-, yellow-, and green- bin algorithm. Overall, there was no significant difference in 17-item Hamilton Rating Scale for Depression (HAMD-17) scores between metabolizer categories based on single-gene phenotyping. Based on the GeneSight categories, however, yellow- and green-bin patients had significantly greater improvement in depression scores than red-bin patients. The authors observed similar trends for total health care visits, medical visits, and disability claims. Results are summarized in Appendix 1.
While these studies provide examples of how GeneSight may be able to predict patient outcomes and health care utilization, it remains uncertain whether using GeneSight to guide medication selection leads to better patient outcomes.
Regulatory Information
The focus of this systematic review was the Assurex GeneSight Psychotropic test. This test is currently not available through the Ontario government. It does not require Health Canada approval; most laboratory tests are subject to approvals at the provincial level.
In the United States, GeneSight Psychotropic is covered by some health insurance plans, including Medicare and Medicaid. According to guidelines from the Centers for Medicare & Medicaid Services, GeneSight may be ordered only by psychiatrists for patients diagnosed with major depressive disorder whose symptoms are refractory after at least one prior neuropsychiatric medication and who continue to experience moderate to severe depression as defined by the Hamilton Rating Scale for Depression.22
Context
The clinical guideline put out by the Clinical Pharmacogenetic Implementation Consortium (CPIC) lists guidance for assessing two of the genes included in the GeneSight test to support the dosing of SSRIs.23 As well, some jurisdictions include information about pharmacogenomics on drug labelling requirements,14 including Health Canada/Santé Canada, the United States Food and Drug Administration (FDA), the European Medicines Agency (EMA), and the Pharmaceutical and Medical Devices Agency, Japan (PMDA). PharmGKB summarizes these labels on its website, organized by medication and available at https://www.pharmgkb.org/view/drug-labels.do. In Ontario, there is no standard method for pharmacogenomic testing at present.
Research Questions
Compared with usual care, what is the effect on depression outcomes of using the GeneSight Psychotropic test to guide the selection of psychotropic medications for patients with mood disorders, anxiety, or schizophrenia?
CLINICAL EVIDENCE REVIEW
Objective
The objective of this study was to assess the effect of the GeneSight Psychotropic test compared with usual care in supporting the selection of psychotropic medications for patients with mood disorders, anxiety, or schizophrenia, within the context of the Ontario Ministry of Health and Long-Term Care.
Methods
Research questions are developed by Health Quality Ontario in consultation with experts, end users, and/or applicants in the topic area.
Sources
We performed a literature search on February 22, 2016, using all Ovid MEDLINE, Embase, PsycINFO, Cochrane Database of Systematic Reviews, Database of Abstracts of Reviews of Effects, CRD Health Technology Assessment Database, Cochrane Central Register of Controlled Trials, and NHS Economic Evaluation Database, for studies published from inception to February 22, 2016.
Search strategies were developed by medical librarians using controlled vocabulary (e.g., Medical Subject Headings) and relevant keywords. The final search strategy was peer-reviewed using the PRESS Checklist (McGowan, 2016). Database auto-alerts were created in MEDLINE, Embase, and PsycINFO, and monitored for the duration of the HTA review. See Appendix 2 for full details, including all search terms. We supplemented searches by conducting general web searches (e.g., Google Scholar), hand-searching bibliographies of identified publications, and by consulting experts.
Literature Screening
A single reviewer reviewed the abstracts and, for those studies meeting the eligibility criteria, we obtained full-text articles.
Inclusion Criteria
English-language full-text publications
Studies published up to February 22, 2016
Studies examining the use of Assurex GeneSight Psychotropic to guide psychotropic medication prescribing among patients with mood disorders, anxiety, or schizophrenia compared with usual (unguided) care
Exclusion Criteria
Studies examining genetic panels other than the GeneSight Psychotropic test
Studies examining the predictive value of a single gene associated with a disorder (e.g., studies looking at candidate genes for association with a phenotype)
Studies examining panels developed for general medication metabolism assessment, not specific to psychotropic medication selection
Animal and in vitro studies
Editorials, case reports, commentaries, or conference abstracts
Outcomes of Interest
-
Primary outcomes (patient outcomes)
∘ Prevention of suicide
∘ Remission of depression symptoms
∘ Response to depression therapy
∘ Depression score
∘ Quality of life
-
Secondary outcomes
∘ Impact on therapeutic decisions
∘ Patient and clinician satisfaction
Data Extraction
We extracted relevant data on study characteristics, risk of bias items, and population, intervention, comparison, outcome, and time (PICOT), collecting information about:
Source (i.e., citation information, contact details, study type)
Methods (i.e., study design, study duration and years, participant allocation, allocation sequence concealment, blinding, reporting of missing data, reporting of outcomes, and whether or not the study compared two or more groups)
Outcomes (i.e., outcomes measured, number of participants for each outcome, outcome definition, and source of information)
Statistical Analysis
We conducted analyses to compare the intervention group (GeneSight Psychotropic) and control groups using Review Manager version 5.3.24 We assumed statistical significance when P < .05. We expressed pooled results as mean differences for continuous data and odds ratios for categorical data. We used a fixed-effects model where there was low between-study heterogeneity based on the interventions and populations described, an I2 ≤30%, or both.25 Where fixed-effects models were inappropriate, we applied a random-effects model. Where pooling of data was not appropriate after considering study design, inclusion criteria, and other sources of heterogeneity between individual studies, we summarized data narratively in evidence tables.
Quality of Evidence
The quality of the body of evidence for each outcome was examined according to the Grading of Recommendations Assessment, Development, and Evaluation (GRADE) Working Group criteria.26 The overall quality was determined to be high, moderate, low, or very low using a step-wise, structural methodology. Details of the GRADE analysis are provided in Appendix 3.
Expert Consultation
In the spring of 2016, we solicited expert consultation on the appropriate use of pharmacogenomic testing to support psychotropic medication selection. Members of the consultation included physicians in the specialty areas of psychiatry and pharmacogenomics. The role of the expert advisors was to provide insight during the development of the research question and contextualize the evidence, providing advice on the potential use of a pharmacogenomic test to support psychotropic medication selection in Ontario. However, the statements, conclusions, and views expressed in this report do not necessarily represent the views of the consulted experts.
Results
Literature Search
The database search yielded 1,807 citations published up to February 22, 2016. After removing duplicates, we reviewed titles and abstracts to identify potentially relevant articles. We obtained the full texts of these articles for further assessment. Four studies met the inclusion criteria.27–31 One was identified through hand-searching of the reference lists of the included studies, along with health technology assessment websites and other sources, to identify additional relevant studies.30
Figure 1 presents the flow diagram for the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA).
Figure 1: PRISMA Flow Diagram.
Abbreviation: PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-analyses.
Source: Adapted from Moher et al.32
Summary of Included Studies
Four studies met the inclusion criteria. Table 2 summarizes the study designs and conclusions of the included studies.
Table 2:
Summary of Studies Examining GeneSight Pharmacogenomic Testing
| Author, Year | Study Design | Study Conclusion |
|---|---|---|
| Hall-Flavin et al, 201228 | Nonrandomized, open-label, consecutive comparative cohort study comparing unguided treatment as usual and guided care with combinatorial genomic testing using GeneSight | Patients in the group guided by GeneSight had greater reduction in depression scores than patients who received unguided treatment |
| Hall-Flavin et al, 201327 | Nonrandomized, open-label, consecutive comparative cohort study comparing unguided treatment as usual and guided care with combinatorial genomic testing using GeneSight | Patients had improved depression outcomes when GeneSight was used for pharmacogenomic testing |
| Winner et al, 201329 | Double-blind randomized controlled trial comparing unguided treatment as usual and guided care with combinatorial genomic testing using GeneSight | Patients who received treatment guided by GeneSight were more likely to respond to therapy, and patients with severe gene-drug interactions who were switched to a more gene-suitable medication had the greatest improvement in depressive symptoms |
| Winner et al, 201530 | Controlled, propensity-matched, prospective cohort study evaluating medication regimens between patients who received guided care with GeneSight vs. usual care, with a focus on cost comparison | Combinatorial testing improved adherence and led to cost savings |
Table 3 outlines the details of the included study populations and interventions. All studies were conducted in the United States, and all studies had a conflict of interest, with authors employed by Assurex, the manufacturer of GeneSight.
Table 3:
Design and Methodology of Studies Examining GeneSight Pharmacogenomic Testing
| Author, Year | Setting | Population | Guided/ Unguided, N | Method | Interventiona (Genes Examined) | Length of Follow-up |
|---|---|---|---|---|---|---|
| Hall-Flavin et al, 201228 | Outpatient behavioural health clinic in St. Paul, MN | Psychiatric patients with a primary diagnosis of major depressive disorder, a HAMD-17 score ≥14, 25–75 years old, and taking at least one of the medications listed by the GeneSight panel. Excluded patients with bipolar disorder (any type), schizophrenia, or schizoaffective disorder | 26/25 | Patients were consecutively screened by their treating physician for eligibility, and allocated in a nonrandomized fashion to the treatment group (results from the GeneSight test were provided to their treating clinician before the start of their therapy to support medication selection) or the unguided control group (treatment as usual) | 5 genes: CYP2D6, CYP2C19, CYP1A2, SLC6A4, 5HTR2A | 8 weeks |
| Hall-Flavin et al, 201327 | Outpatient psychiatry unit at a hospital in La Crosse, WI | Psychiatric patients with a primary diagnosis of major depressive disorder or depressive disorder not otherwise specified, a HAMD-17 score ≥14, and 18–72 years old. Excluded patients with bipolar disorder type I, schizophrenia, or schizoaffective disorder | 114/113 | The first of the two consecutive groups received treatment as usual. The second group's treating clinicians were given GeneSight results to support medication selection. Physicians were not given additional education or training on pharmacogenomics or GeneSight | 5 genes: CYP2D6, CYP2C19, CYP1A2, SLC6A4, 5HTR2A | 8 weeks |
| Winner et al, 201329 | Outpatient clinic at a mental health services clinic in Grand Rapids, MI | Patients with a diagnosis of major depressive disorder or depressive disorder not otherwise specified, and a HAMD-17 score ≥14. Excluded patients with bipolar disorder (any type), schizophrenia, schizoaffective disorder, or substance abuse or dependence | 26/25 | Patients were randomized to the intervention group (treating psychiatrist or psychiatric nurse practitioner received the GeneSight test results within 2 days of enrolment) or the control group (treatment as usual). Analysts and patients were blinded to their treatment group allocation | 6 genes: CYP2D6, CYP2C19, CYP2C9, CYP1A2, SLC6A4, 5HTR2A | 10 weeks |
| Winner et al, 201530 | Various, United States | Psychiatric patients diagnosed with anxiety, depressive disorders, bipolar disorders, or other psychotic disorders, and prescribed one of the 26 antidepressant or antipsychotic medications covered by GeneSight at the time. Patients were eligible if they had no prescriptions in the 180 days prior to the eligible GeneSight medication prescription, maintained continuous pharmaceutical support in the previous 180 days, or experienced an augmentation in prescribed medication or dosage in the previous 90 days | 2,168/10,880 | Clinicians of eligible patients were contacted and asked if they authorized GeneSight testing. Patients were then propensity-matched 1:5 on date of project enrolment, age, sex, psychotropic medication, and primary diagnosis against controls from a large administrative data set of 65 million individuals from the participating health insurance plan who did not receive testing. Patients were then followed for 1 year using administrative data | Specific version not reportedb | 1 year |
Abbreviations: HAMD-17, 17-item Hamilton Rating Scale for Depression.
See Appendix 1 for more details about the specific alleles.
While the specific version of GeneSight was not reported in the study, the reported enrolment period was from September 2011 to December 2013. Winner et al13 published a paper that reported the six-gene version of GeneSight was used in April 2011, and we found a press release dated July 2013 announcing a new version of GeneSight that included a new variant,33 leading us to believe that the six-gene version was used for the majority of this study.
Three of the studies limited the populations to patients with a diagnosis of major depressive disorder or depressive disorder not otherwise specified (treated by psychiatrists), and they excluded patients with other diagnoses such as bipolar type I, schizophrenia, or schizoaffective disorder.27–29 One study was broader in its inclusion criteria and included all psychiatric patients diagnosed with depressive disorder, anxiety, bipolar disorder, or other psychiatric disorders. As well, this study did not explicitly limit the ordering of GeneSight to psychiatrists.30
All studies used the same protocol to identify polymorphisms. Genotyping for CYP2D6, CYP2C19, and CYP1A2 (plus CYP2C9 in Winner et al 2013b13) was done using the Luminex xTAG Multiplex Technology (Luminex, Austin, TX) with polymerase chain reaction (PCR) amplification of the relevant regions and using Exonuclease I and Shrimp Alkaline Phosphatase to clarify. Serotonin transporter and receptor genes SLC6A4 and HTR2A were amplified using PCR. The restriction enzyme MSPO (Moraxella species) was used on HTR2A, and both SLC6A4 and HTR2A were run on a 2% gel to genotype.
Results for Primary Outcomes
The primary outcomes of interest were patient outcomes: prevention of suicide, remission of depression symptoms, response to depression therapy, depression score, and quality of life.
Three depression scores were used across the included studies. The HAMD-17 is administered by health care professionals to patients who have already been diagnosed with depression to assess severity; a higher score indicates increased severity of depression.34 The 16-item Quick Inventory of Depressive Symptomatology is also a depression severity scale with a clinician-rated component (QIDS-C16) and a patient self-rated component (QIDS-SR), both of which have demonstrated internal consistency and are commonly used in practice as valid measures of depression.35 The 9-item Patient Health Questionnaire (PHQ-9) is a validated survey completed by patients to diagnose major depressive disorder or other depressive syndromes; a higher score indicates increased severity of depressive symptoms.36
Prevention of Suicide
When managing patients with depression, prevention of suicide is the most important outcome, as advised by experts consulted for this review. However, none of the included studies reported on suicide.
Remission of Depression Symptoms
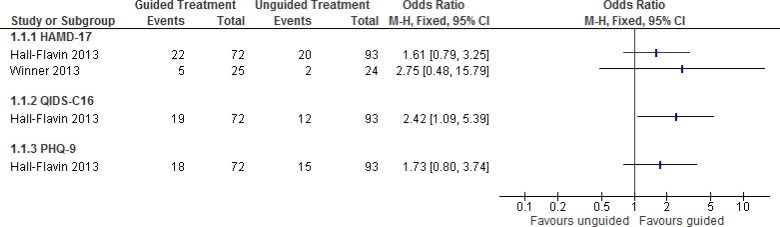
Total remission is the primary goal when treating patients with depression, as advised by the experts consulted for this review. Figure 2 summarizes results from the two studies that reported on patient remission. In both studies, patients were considered to be in remission when they achieved HAMD-17 scores <8, QIDS-C16 scores <5, and PHQ-9 scores <5.27,29
Figure 2: Proportion of Patients With Remission of Depression Symptomsa,b.
Abbreviations: CI, confidence interval; HAMD-17, 17-item Hamilton Rating Scale for Depression; M-H, Mantel-Haenszel; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating.
aHall-Flavin et al reported nonsignificant results for HAMD-17 and PHQ-9, but calculations were estimates based on raw data presented in the original report. For HAMD-17, there were some rounding differences in the data presented here compared with the original study.27
bHall-Flavin et al had a follow-up period of 8 weeks; Winner et al had a follow-up period of 10 weeks.27,29
One study, Hall-Flavin et al,27 also conducted sensitivity analyses using the intention-to-treat analytical strategies of expected maximum and last observation carried forward; they found results to be consistent and/or stronger in favour of guided therapy than those reported here. There was a statistically significant benefit in depression remission for GeneSight compared with usual treatment when depressive symptoms were assessed using QIDS-C16, but no significant difference when using HAMD-17 or PHQ-9. This was based on very low quality evidence (Table 4), as the findings were inconsistent with respect to the effectiveness of GeneSight to improve remission from depression, and they depended on the scale used to measure depression (Figure 2).
Table 4:
GRADE Evidence Profile for Remission of Depression Symptoms
| Number of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| HAMD-17 | |||||||
| 1 (RCT) 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| QIDS-C16 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| PHQ-9 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAMD-17, 17-item Hamilton Rating Scale for Depression; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating; RCT, randomized controlled trial.
Risk of bias assessment details are available in Appendix 3.
Wide confidence intervals impacted our confidence in the estimate. Given the small number of events, the findings may not be as robust, which leads us to have some uncertainty about the results observed. In particular, the relatively large treatment effect given the small sample size of the RCT may be due to prognostic imbalance and warrants caution in interpretation of the results.
Response to Depression Therapy
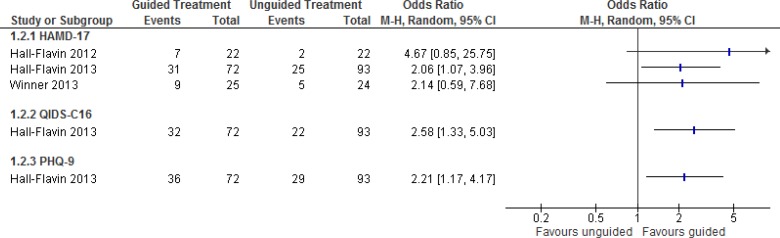
Three studies reported the proportion of patients who responded to depression therapy.27–29,31 In these studies, responders were defined as having achieved at least a 50% reduction in depression scores.27–29,31
Figure 3 analyzes the results for response rate. Hall-Flavin et al28 did not report response rates in their publication, but data for that population were available from a cost-effectiveness analysis published by Hornberger et al.31
Figure 3: Proportion of Patients With Response to Depression Therapya,b,c.
Abbreviations: CI, confidence interval; HAMD-17, 17-item Hamilton Rating Scale for Depression; M-H, Mantel-Haenszel; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating.
aFor Hall-Flavin et al, the numbers presented here present some rounding differences from the study, which reported an odds ratio for the HAMD-17 score of 2.06 (95% CI, 1.07–3.95).27
bThe response rate for Hall-Flavin et al28 was not reported in the original study, but data were available from a cost-effectiveness analysis published by Hornberger et al.31
cHall-Flavin et al had a follow-up period of 8 weeks; Winner et al 2013 had a follow-up period of 10 weeks.27,29
One study, Hall-Flavin et al27 conducted sensitivity analyses using the intention-to-treat analytical strategies of expected maximum and last observation carried forward and found that results were consistent and/or stronger in favour of guided therapy than those reported here. A significantly greater proportion of patients had a response to depression therapy with GeneSight-guided care than those who received usual care (Figure 3), based on low to very low quality evidence (Table 5).
Table 5:
GRADE Evidence Profile for Response to Depression Therapy
| Number of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| HAMD-17 | |||||||
| 1 (RCT) 2 (observational) | Serious limitationsa | No limitations | No limitations | No limitations | None detected | None | ⊕⊕ Low |
| QIDS-C16 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| PHQ-9 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAMD-17, 17-item Hamilton Rating Scale for Depression; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating; RCT, randomized controlled trial.
Risk of bias assessment details are available in Appendix 3.
Wide confidence intervals impacted our confidence in the estimate of effect.
Depression Score
Across the studies that reported on changes in depression scores, patients in the guided and unguided study groups were considered to be similar at baseline.27–29
Table 6 summarizes the effect on measures of depression among patients who received guided care compared with those who did not (at end-of-study follow-up). Overall, patients who received guided care with GeneSight had a greater reduction in depression than those who had treatment as usual.
Table 6:
Depression Score at End of Study
| Author, Year | Length of Follow-up | % Reduction in Score (Improvement in Depression) | Significancea | |
|---|---|---|---|---|
| Guided | Unguided | |||
| HAMD-17 | ||||
| Hall-Flavin et al, 201228 | 8 weeks | 30.8% | 18.2% | P = .04; adjusted P = .05 |
| Hall-Flavin et al, 201327 | 8 weeks | 46.9% | 29.9% | P < .0001; adjusted P < .0001 |
| Winner et al, 201329 | 10 weeks | 30.8% | 20.7% | P = .28 |
| QIDS-C16 | ||||
| Hall-Flavin et al, 201228 | 8 weeks | 31.2% | 7.2% | P = .002; adjusted P = .003 |
| Hall-Flavin et al, 201327 | 8 weeks | 44.8% | 26.4% | P < .0001; adjusted P < .0001 |
| Winner et al, 201329 | 10 weeks | 27.6% | 22.1% | NS |
| PHQ-9 | ||||
| Hall-Flavin et al, 201327 | 8 weeks | 40.1% | 19.5% | P < .0001; adjusted P = .002 |
| Winner et al, 201329 | 10 weeks | 35.4% | 21.3% | P = .18 |
Abbreviations: ANOVA, analysis of variance; HAMD-17, 17-item Hamilton Rating Scale for Depression; NS, not significant; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating.
A repeated-measures ANOVA mixed-model approach was used, adjusting for time (in weeks) and treatment group.
No statistically significant differences were observed between groups at 2 weeks except for Hall-Flavin et al,27 who observed a significantly greater depression score with the QIDS-C16 measure at baseline in the guided group than in the unguided group (17.5 vs. 16.0, P = .003). At 4 weeks, Hall-Flavin et al27 found a significantly greater improvement in the guided group on the HAMD-17 (P = .0002) and QIDS-C16 (P = .0002), but not on the PHQ-9 (P = NS). At 6 weeks, Winner et al29 saw similarly greater improvement on the HAMD-17 score in the guided group (35.4% improvement vs. 18.5% improvement in the unguided group, P = .04). Sensitivity analyses conducted by the studies using expected maximum and last observation carried forward to account for missing data demonstrated that results were robust and conclusions remained aligned to the primary analyses summarized in Table 6.
Based on very low quality evidence (Table 7), improvements in depression score were greater among patients who received GeneSight-guided care than those who received usual care.
Table 7:
GRADE Evidence Profile for Depression Score
| Number of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| HAMD-17 | |||||||
| 1 (RCT) 2 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| QIDS-C16 | |||||||
| 2 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| PHQ-9 | |||||||
| 2 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAMD-17, 17-item Hamilton Rating Scale for Depression; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating.
Risk of bias assessment details are available in Appendix 3.
Wide confidence intervals impacted our confidence in the estimate of effect; experts advised that a clinically meaningful difference in HAMD-17 was two to three points.
Subgroup Analyses
Some studies conducted subgroup analyses based on patients' bin status. Table 8 summarizes the results comparing the end-of-study reduction in depression score among those who received guided versus unguided care in patients who took red-bin medications at the start of each study.
Table 8:
Depression Score Among Patients Who Took Red-Bin Medications
| Author, Year | Measure | % Reduction in Score (Improvement in Depression) | Significance | |
|---|---|---|---|---|
| Guided | Unguided | |||
| Hall-Flavin et al, 201327 | HAMD-17 | 42.5% | 16.6% | P = .01 |
| QIDS-C16 | 41.9% | 11.0% | P = .004 | |
| Winner et al, 201329 | HAMD-17 | 32%a | 0.8% | P = .06 |
Abbreviations: HAMD-17, 17-item Hamilton Rating Scale for Depression; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating.
Percentage reduction is an estimate based on Figure 4 published in the study.29
The authors also observed a difference among patients in the unguided care group who were prescribed red-bin medications compared with those who received yellow- or green-bin medications. Patients who were prescribed red-bin medications but received unguided care demonstrated an improvement in HAMD-17 and QIDS-C16 scores of 0.8% to 16.6%; patients on yellow- and green-bin medications improved significantly more, from 26.5% to 36.1% (P = .007).27,29
Quality of Life
None of the studies reported on quality of life.
Results for Secondary Outcomes
Impact on Therapeutic Decisions
We considered changes to medication regimens as a measure of therapeutic decisions, presuming that modifications indicated that patients were not responding to depression therapy or had had adverse reactions that prompted decisions to change the regimen. All included studies reported some measure of medication change (Table 9).
Table 9:
Impact on Therapeutic Decisions—Modifications to Medication Regimens, Adherence to Medication Regimens, and Polypharmacy
| Author, Year | Measure | Results | Significance | |
|---|---|---|---|---|
| Guided | Unguided | |||
| Modifications | ||||
| Hall-Flavin et al, 201327 | Proportion of patients who had a changea in their medication from baseline | 76.8% | 44.1% | P < .0001 |
| Winner et al, 201329 | Proportion of patients who had a changea in their medication from baseline to end of study | 53% | 58% | P = .66 |
| Adherence | ||||
| Winner et al, 201530 | Adherenceb at end of follow-up | 0.74 | 0.79 | P < .0001 |
| Change from baseline in adherenceb | 0.111 | −0.01 | P < .0001 | |
| Differences between rates of medication discontinuation before and after the start of study | −7.6% | +0.3% | P < .0001 | |
| Mean time to discontinuation of the initial medication from the start of study | 103 days | 134 days | P < .0001 | |
| Polypharmacy | ||||
| Hall-Flavin et al, 201228 | Difference in mean number of medications per patient at the end of the study compared with the beginning | −2.7 (SD 3.5)c | −2.2 (SD 3.4)c | Difference of the means: 0.5 (SD 6.7)c |
| Winner et al, 201329 | Mean number of psychiatric medications per patient at end of study | 1.9 | 1.7 | P = .27 |
| Winner et al, 201530 | Increase in average number of medications taken from baseline to end of follow-up | 0.88 | 1.07 | P < .0001 |
Abbreviation: SD, standard deviation.
Change included a switch of medication or augmentation in dosage.
Adherence was the ratio of the proportion of days on a medication over the total covered days from the index prescription, with discontinuation defined as ≥45 days between refills.
Calculated based on data provided in the original study; estimate around the variance is conservative given that within-study correlation was not accounted for. Reported mean medications at baseline in the guided and unguided groups were 4.4 (SD 3.4) and 4.4 (SD 3.13), respectively. Reported mean medications at the end of the study in the guided and unguided groups were 1.7 (SD 0.84) and 2.2 (SD 1.4), respectively.28
One study found a significant difference in the proportion of patients who discontinued the medication prescribed at the start of the study between the guided group (60.9%) and the unguided group (40.9%; P < .001).30 However, patients who received guided care were also more likely to discontinue their medication during the study period (53.3%) than those who received treatment as usual (41.2%; P < .001).30 The guided group also had a slightly shorter but statistically significant (P < .001) time before discontinuing the initial medication, at 150 days after the start of the study compared with 152 days for the unguided group.30
Based on very low quality evidence (Table 10), the findings were inconsistent as to whether GeneSight-guided care improved therapeutic decisions compared with usual care (Table 9).
Table 10:
GRADE Evidence Profile for Impact on Therapeutic Decisions
| Number of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| 1 (RCT) 3 (observational) | Serious limitationsa | Serious limitationsb | Very serious limitationsc | No limitations | None detected | None | ⊕ Very low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; RCT, randomized controlled trial.
Risk of bias assessment details are available in Appendix 3.
Results were inconsistent between the studies: some demonstrated an increase in the mean number of medications per patient by end of study, and others a decrease.
Outcomes were indirect measures of the outcome of interest (the impact of guided therapy on therapeutic decision-making).
Subgroup Analyses
Table 11 summarizes the subgroup analyses conducted by studies that assessed results based on whether patients were prescribed green-bin, yellow-bin, or red-bin medications according to the GeneSight test.
Table 11:
Modifications to Medication Regimens by Subgroup Based on GeneSight Results
| Author, Year | Measure | Results | Significance | |
|---|---|---|---|---|
| Guided | Unguided | |||
| Hall-Flavin et al, 201228 | Proportion of patients taking a green-bin medication at end of study | 71%a | 21%a | NR |
| Proportion of patients taking a yellow-bin medication at end of study | 23%a | 57%a | NR | |
| Proportion of patients taking a red-bin medication at end of study | 5.9% | 21.4% | P = .02 | |
| Hall-Flavin et al, 201327 | Proportion of patients taking a green-bin medication at end of study | 40% | 27.6% | NSb |
| Absolute increase in percentage points of the proportion of patients taking a green-bin medication compared with baseline | 13.4 | 1.7 | NSb | |
| Proportion of patients taking a red-bin medication who changed their medication by end of study | 93.8% | 55.6% | P = .01 | |
| Winner et al, 201329 | Proportion of patients taking a red-bin medication at baseline | 100% | 50% | P = .02 |
| Winner et al, 201530 | Change in proportion of patients taking a green-bin medication from start of study to end of studyc | +13.3% | NR | NR |
| Change in proportion of patients taking a yellow-bin medication from start of study to end of studyc | −4.8% | NR | NR | |
| Change in proportion of patients taking a red-bin medication from start of study to end of studyc | −8.5% | NR | NR | |
Abbreviation: NR, not reported.
Estimates based on visual inspection of a graph reported in the study.
Study reported as not significant, but did not provide detailed statistics.
Within the previous 90 days.
By the end of the studies, most patients were taking green-bin or yellow-bin medications, and few were taking red-bin medications.
Patient and Clinician Satisfaction
One study reported on satisfaction. The authors asked clinicians to rate their level of satisfaction, and to estimate their patients' level of satisfaction (Table 12).
Table 12:
Satisfaction
| Author, Year | Measure | Intervention/Control, N | Results | Significance | |
|---|---|---|---|---|---|
| Guided | Unguided | ||||
| Patient Satisfaction | |||||
| Hall-Flavin et al, 201327 | Physician perception of patient satisfaction; proportion rated as having very high perceived satisfaction | 37/88 | 40.5% | 14.8% | P = .008 |
| Clinician Satisfaction | |||||
| Hall-Flavin et al, 201327 | Physician satisfaction with care | 37/88 | 94.6% | 61.8% | P = .0007 |
| Physician confidence in choice of medication | 37/88 | 91.9% | 61.8% | P = .003 | |
Based on very low quality evidence (Table 13), there was significantly greater patient satisfaction among those who received GeneSight-guided care than among those who received usual care (Table 12). Similarly, based on low quality evidence (Table 13), there was greater clinician satisfaction with GeneSight-guided care than with usual care (Table 12).
Table 13:
GRADE Evidence Profile for Satisfaction
| Number of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Patient Satisfaction | |||||||
| 1 (observational) | Serious limitationsa | No serious limitations | Very serious limitationsb | No serious limitations | None detected | None | ⊕ Very low |
| Clinician Satisfaction | |||||||
| 1 (observational) | Serious limitationsa | No serious limitations | No serious limitations | No serious limitations | None detected | None | ⊕⊕ Low |
Abbreviation: GRADE, Grading of Recommendations Assessment, Development, and Evaluation.
Risk of bias assessment details are available in Appendix 3.
Measure of patient satisfaction was based on clinician perception of their patients' satisfaction.
Discussion
We identified one systematic review on this topic. It was published by authors affiliated with the manufacturer, and it identified the same studies included in this review and drew similar conclusions to this report.37
Limitations
There are multiple versions of the GeneSight Psychotropic test. The results from the studies were based on an algorithm that used a combination of five or six genes and may not be generalizable to a version of GeneSight containing eight, 12, or more genes. As well, the three studies that reported our primary outcomes of interest limited their design to psychiatrists; we cannot draw conclusions about the effectiveness of the test when used by other clinicians. One study by the manufacturers found that non-psychiatrist primary care physicians followed the recommendations of GeneSight more closely than psychiatrists, but this study did not demonstrate whether this difference had an impact on patient outcomes, so it did not meet the inclusion criteria for this review.30 Similarly, GeneSight has been proposed for use in patients with mood disorders, anxiety, or schizophrenia, but the three studies that reported results for our primary outcomes of interest limited their populations to patients with major depressive disorder, excluding patients with anxiety alone without depression, patients with bipolar disorder, or schizophrenia. Taken together, these factors limit the generalizability of the body of evidence. Looking to other jurisdictions, at the time of writing this report, the United States Centers for Medicare & Medicaid Services covered GeneSight only if it was ordered by psychiatrists for patients with major depressive disorder whose symptoms were refractory after at least one prior neuropsychiatric medication and who continued to have moderate to severe depression as defined by the Hamilton Rating Scale for Depression.22
A second limitation was the uncertainty around the level of evidence supporting each medication included in the GeneSight report. Each medication has its own evidence profile, but the quality of the evidence supporting green-, yellow-, or red-bin classifications in GeneSight is not clear in the current version of the GeneSight report as clinicians would not know the strength of the evidence behind what moves a medication from green bin- to another status. Assurex has indicated that a revised version of the report, expected in early 2017, will be more transparent. The updated version is expected list which genes are related to which medications.
Comparable Tests
This review focused on the Ontario context, which at the time of review did not cover pharmacogenomic testing for patients with mood disorders. However, this test, and others similar to GeneSight are available if paid for by the patient. The list in Table 14 is not exhaustive or comprehensive—simply a summary of known tests similar in purpose to GeneSight Psychotropic providing a pharmacogenomic panel to support psychotropic medication selection.
Table 14:
Comparable Pharmacogenomic Panels to Support Psychotropic Medication Selection
| Company | Assurex | CNSdose | CRL Corp | Genele X | Genomind | MDL-Labs |
|---|---|---|---|---|---|---|
| Test | GeneSight | CNSDose | SureGene | You Script psychotropic plus | Genecept | Psychiatric pharmacogenetic |
| Websitea | https://genesight.com/ | https://www.cnsdose.com/ | http://www.crlcorp.com/services/personalized-medicine/tests/ | http://genelex.com/ | https://genomind.com/ | http://www.mdllabs.com/providers/tests/psychiatric-pharmacogenetic-test-panel |
| Number of genes | 8b | 3c | 19 | 12 | 19 | 4 |
| List of genes | ABC transportersc | |||||
| ADRA2A | ADRA2A | |||||
| ANK3 | ||||||
| BDNF | ||||||
| CACNA1C | ||||||
| COMT | COMT | COMT | ||||
| CYP1A2 | CYP1A2 | CYP1A2 | CYP1A2 | |||
| CYP2B6 | CYP2B6 | CYP2B6 | CYP2B6 | |||
| CYP2C19 | CYP2C19 | CYP2C19 | CYP2C19 | CYP2C19 | ||
| CYP2C9 | CYP2C9 | CYP2C9 | CYP2C9 | |||
| CYP2D6 | CYP2D6 | CYP2D6 | CYP2D6 | CYP2D6 | ||
| CYP3A4 | CYP3A4 | CYP3A4 | CYP3A4 | CYP3A | ||
| CYP3A5 | CYP3A5 | |||||
| CYP450c | DRD2 | |||||
| FOLH1 | ||||||
| GLP1R | GRIK4 | GRIK1 | ||||
| HLA-A | ||||||
| HTR2A | 5HTR2A | HTR2A | ||||
| 5HTR2C | HTR2C | 5HT2C | ||||
| MC4R | ||||||
| MTHFR | MTHFR | MTHFR | ||||
| OPRM1 | OPRM1 | |||||
| SLC6A4 | SLC6A4 | SLC6A4/5-HTT | SLC6A4 | |||
| SULT4A1 | ||||||
| UGT1A1 | ||||||
| VKORC1 |
All websites were accessed on May 2, 2016.
Included genes were based on the application received from Assurex for review.
Specific genes in these gene groups were not identified on the company website, or in the academic paper by Singh et al.38 We are aware of two other similar tests, but we were unable to locate specific details about these tests at the time of this review. These tests are psynome2 by psynomics and PsychINDx by curidium medica.
Conclusions
Patients who received guided medication selection with the GeneSight Psychotropic test demonstrated improved response to depression therapy and greater improvement in measures of depression and patient and clinician satisfaction than patients who received treatment as usual. However, there were no observed differences in rates of complete remission from depression. These findings are uncertain because they are supported by low to very low confidence in the evidence (Table 15).
Table 15:
Summary of Results for Review of GeneSight to Support Psychotropic Medication Selection Among Patients With Mood Disorders
| Outcome | Measure | Results | GRADE |
|---|---|---|---|
| Patient Outcomes | |||
| Suicide | Mortality | None reported | NA |
| Remission of depression symptoms | Depression below a predefined threshold: HAMD-17 <8, QIDS-C16 <5 or PHQ-9 <5 | No significant difference between groupsa | Very low |
| Response to depression therapy | At least 50% reduction in depression scores based on HAMD-17, QIDS-C16, or PHQ-9 | Favoured guided therapy | Low to very low |
| Measures of depression | % reduction in score (improvement in depression) HAMD-17 | Favoured guided therapy | Very low |
| QIDS-C16 | Favoured guided therapy | Very low | |
| PHQ-9 | Favoured guided therapy | Very low | |
| Quality of life | Validated measures of quality of life | None reported | NA |
| Secondary Outcomes | |||
| Impact on therapeutic decisions | Changes to medications | Inconsistent measurement outcome resulted in either no significant difference or observed difference in favour of guided therapy | Very low |
| Satisfaction | Patient satisfaction | Favoured guided therapy | Very low |
| Clinician satisfaction | Favoured guided therapy | Low | |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAMD-17, 17-item Hamilton Rating Scale for Depression; NA, not applicable; NS, not significant; PHQ-9, 9-item Patient Health Questionnaire; QIDS-C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating.
No significant difference when using the HAMD-17 or PHQ-9 measures; a significant result was observed with the QIDS-C16 in favour of guided care.
Acknowledgments
The medical editor was Jeanne McKane; others involved in the development and production of this report were Claude Soulodre, Kellee Kaulback, Andree Mitchell, Anil Thota, Nancy Sikich, and Irfan Dhalla.
We are grateful to the following for sharing their expertise in the areas of psychiatry and pharmacogenomics.
Dr. Roger McIntyre, Head, Mood Disorders Psychopharmacology Unit at the University Health Network; Professor of Psychiatry and Pharmacology, University of Toronto; Executive Director, Brain and Cognition Discovery Foundation (BCDF).
Dr. Richard Kim, Professor of Medicine, Physiology & Pharmacology, and Oncology, Wolfe Medical Research Chair in Pharmacogenomics, Department of Medicine, Schulich School of Medicine & Dentistry, University of Western Ontario.
We are grateful to Assurex Health for providing technical information about the GeneSight Psychotropic test.
Glossary
LIST OF ABBREVIATIONS
- CPIC
Clinical Pharmacogenetic Consortium
- EMA
European Medicines Agency
- FDA
Food and Drug Administration
- GRADE
Grading of Recommendations Assessment, Development, and Evaluation
- HAMD-17
17-item Hamilton Rating Scale for Depression
- LHIN
Local Health Integration Network
- PHQ-9
9-item Patient Health Questionnaire
- PICOT
Population, intervention, comparison, outcome and time
- PMDA
Pharmaceutical and Medical Devices Agency, Japan
- QIDS-C16
16-item Quick Inventory of Depression Symptomatology, clinician rating
- SSRI
Selective serotonin reuptake inhibitor
Glossary
GLOSSARY
- Fixed effects
A meta-analysis method (a study of studies) that takes into account only the variation in effects among the individual participants across all studies. It does not look at the variation in results among the studies. This analysis assumes differences in study methods do not affect the overall outcome.
- Gene
A specific segment of a chromosome (a DNA strand) that is responsible for passing on a specific trait through inheritance.
- Genome
The entire genetic makeup (all DNA) that includes the genes and non-gene segments of a chromosome.
- Genomics
The field of study that focuses on the genome of an organism (as distinct from genetics, which focuses on the effects of a single gene). It includes how the expression of genes is affected by environmental factors, such as a person's lifestyle.
- Genotype
The genome of a specific person. Genotype may also be used to refer to a segment of a person's genome (usually a gene).
- Pharmacogenomics
A field of study that looks at how a person's genotype may affect their response to a drug.
- Phenotype
The physical characteristics resulting from a person's genetic profile (their genotype).
APPENDICES
Appendix 1: Additional Clinical Evidence
Table A1:
Genes Included in the GeneSight Psychotropic Test
| Gene | Location | Variants Detected by the Test | Mechanism of Action |
|---|---|---|---|
| CYP1A2 | 15q24.1 | 15 SNPs: −163C>A, −246delT, −3860 G>A, −729C>T, −739 T>G, 125C>G, 2116 G>A, 2473 G>A, 2499 A>T, 349 G>A, 3533 G>A, 5090C>T, 5166 G>A, 5347C>T, 558C>A | A member of the cytochrome P450 family of liver enzymes, which plays an important role in drug metabolism |
| CYP2B6 | 19q13.2 | 2 SNPs: A785G (*4) and G516T (*6) | A member of the cytochrome P450 family of liver enzymes, which plays an important role in drug metabolism |
| CYP2C19 | 10q23.33 | Detection of *1, *2, *3, *4, *5, *6, *7, *8 and *17 alleles | A member of the cytochrome P450 family of liver enzymes, which plays an important role in drug metabolism |
| CYP2C9 | 10q23.33 | Detection of *1, *2, *3, *4, *5, and *6 alleles | A member of the cytochrome P450 family of liver enzymes, which plays an important role in drug metabolism |
| CYP2D6 | 22q13.2 | Detection of *1, *2, *2 A, *3, *4, *5 (deletion of allele), *6, *7, *8, *9, *10, *11, *12, *14, *15, *17, and *41 alleles. Multiple copies of the allele | A member of the cytochrome P450 family of liver enzymes, which plays an important role in drug metabolism |
| CYP3A4 | 7q22.1 | Detection of *1, *13, *15A, and *22 alleles | A member of the cytochrome P450 family of liver enzymes, which plays an important role in drug metabolism |
| HTR2A | 13q14.2 | 3 genotypes: A/A, A/G, and G/G | A serotonergic gene where variations affect the mechanism of action for the transporter activity of certain SSRIs (e.g., adverse drug effects) |
| SLC6A4 | 17q11.2 | Detection of three genotypes of two alleles: homozygous for the long allele, homozygous for the short allele, and heterozygous for long and short | A serotonergic gene where variations affect the mechanism of action for the receptor activity of certain SSRIs (e.g., reduced response rate and increased time to response) |
| The long allele is characterized by a 44 base-pair insertion in the promoter region and a 419 base-pair fragment of DNA vs. 375 in the short allele, a visible difference on gel electrophoresis |
Abbreviation: SNP, single nucleotide polymorphism; SSRI, selective serotonin reuptake inhibitor.
Table A2:
Results for Differences Across Patient Phenotypes Identified With Single Genotyping vs. Combinatorial Genomic Testing (GeneSight)a
| Outcome | Patient Gene Group | Unguided Patients, N | Results | |
|---|---|---|---|---|
| Combinatorial Genomics (GeneSight) | Single-Gene Phenotyping | |||
| HAMD-17 improvement | CYP2D6 | 117 | P = .003b | P = .96 |
| CYP2C19 | 80 | P = .04b | P = .52 | |
| CYP1A2 | 35 | P = .03b | P = .33 | |
| SLC6A4 | 66 | P = .27 | P = .43 | |
| HTR2A | 5 | P = .16 | P = .16 | |
| Total health care visits | CYP2D6 | 79 | P = .04b | P = .11 |
| CYP2C19 | 59 | P = .04b | P = .02b | |
| CYP1A2 | 32 | P = .01b | P = .83 | |
| CYP2C9 | 10 | P = .55 | P = .32 | |
| SLC6A4 | 60 | P = .77 | P = .63 | |
| HTR2A | 15 | P = .78 | P = .26 | |
| Medical visits | CYP2D6 | 79 | P = .06 | P = .26 |
| CYP2C19 | 59 | P = .14 | P = .05b | |
| CYP1A2 | 32 | P = .02b | P = .61 | |
| CYP2C9 | 10 | P = .17 | P = .44 | |
| SLC6A4 | 60 | P = .51 | P = .58 | |
| HTR2A | 15 | P = .61 | P = .18 | |
| Disability claims | CYP2D6 | 79 | P = .002b | P = .55 |
| CYP2C19 | 59 | P = .001b | P = .62 | |
| CYP1A2 | 32 | P = .14 | P = .35 | |
| CYP2C9 | 10 | P = .65 | P = .44 | |
| SLC6A4 | 60 | P = .44 | P = .26 | |
| HTR2A | 15 | P = .80 | P = .51 | |
Abbreviations: HAMD-17, 17-item Hamilton Rating Scale for Depression.
Results from Altar et al.16
Statistically significant.
Appendix 2: Literature Search Strategies
Search date: Feb 22, 2016
Librarians: Corinne Holubowich
Databases searched: All Ovid MEDLINE, Embase, PsycINFO, Cochrane Database of Systematic Reviews, Database of Abstracts of Reviews of Effects, CRD Health Technology Assessment Database, Cochrane Central Register of Controlled Trials, and NHS Economic Evaluation Database
Database: EBM Reviews - Cochrane Central Register of Controlled Trials <January 2016>, EBM Reviews - Cochrane Database of Systematic Reviews <2005 to February 19, 2016>, EBM Reviews - Database of Abstracts of Reviews of Effects <1st Quarter 2016>, EBM Reviews - Health Technology Assessment <1st Quarter 2016>, EBM Reviews - NHS Economic Evaluation Database <1st Quarter 2016>, Embase <1980 to 2016 Week 08>, All Ovid MEDLINE(R) <1946 to Present>, PsycINFO <1967 to February Week 2 2016>
Search Strategy:
| 1 | Depressive Disorder, Major/ (26865) |
| 2 | ((depression* or depressive* or melancholia* or paraphrenia* or psychos?s) adj2 (major or disorder* or involution* or unipolar*)).tw. (187247) |
| 3 | Mood Disorders/ (29497) |
| 4 | ((affective or mood) adj disorder*).tw. (90895) |
| 5 | Anxiety Disorders/ (48451) |
| 6 | Bipolar Disorder/ (97611) |
| 7 | ((anxiety adj (disorder* or neuros?s)) or ((bipolar or manic) adj (psychos?s or disorder* or depression* or depressive*))).tw. (165660) |
| 8 | or/1–7 (432557) |
| 9 | Pharmacogenetics/ (25576) |
| 10 | (pharmacogenetic* or pharmacogenomic*).tw. (29773) |
| 11 | Genetic Testing/ (59139) |
| 12 | Genotyping Techniques/ (6016) |
| 13 | (((genetic* or gene or genes) adj2 (test or tests or testing or panel* or assess* or screen* or profil* or algorithm* or combinatorial)) or psychotropic panel*).tw. (204909) |
| 14 | (genotype adj (assignment* or method* or technique*)).tw. (948) |
| 15 | (genesight or assurex).tw. (25) |
| 16 | or/9–15 (287465) |
| 17 | 8 and 16 (3171) |
| 18 | Case Reports/ or Comment.pt. or Editorial.pt. or Letter.pt. or Congresses.pt. (4518636) |
| 19 | 17 not 18 (3096) |
| 20 | exp Animals/ not (exp Animals/ and Humans/) (13325189) |
| 21 | 19 not 20 (2251) |
| 22 | limit 21 to english language [Limit not valid in CDSR,DARE; records were retained] (2150) |
| 23 | 22 use pmoz,cctr,coch,dare,clhta,cleed (974) |
| 24 | major depression/ (141961) |
| 25 | ((depression* or depressive* or melancholia* or paraphrenia* or psychos?s) adj2 (major or disorder* or involution* or unipolar*)).tw. (187247) |
| 26 | mood disorder/ (56353) |
| 27 | ((affective or mood) adj disorder*).tw. (90895) |
| 28 | anxiety disorder/ (92738) |
| 29 | bipolar disorder/ (97611) |
| 30 | ((anxiety adj (disorder* or neuros?s)) or ((bipolar or manic) adj (psychos?s or disorder* or depression* or depressive*))).tw. (165660) |
| 31 | or/24–30 (519047) |
| 32 | pharmacogenomics/ (17736) |
| 33 | (pharmacogenetic* or pharmacogenomic*).tw. (29773) |
| 34 | genetic screening/ (85784) |
| 35 | genotyping technique/ (7207) |
| 36 | (((genetic* or gene or genes) adj2 (test or tests or testing or panel* or assess* or screen* or profil* or algorithm* or combinatorial)) or psychotropic panel*).tw. (204909) |
| 37 | (genotype adj (assignment* or method* or technique*)).tw. (948) |
| 38 | (genesight or assurex).tw. (25) |
| 39 | or/32–38 (299732) |
| 40 | 31 and 39 (3541) |
| 41 | Case Report/ or Comment/ or Editorial/ or Letter/ or conference abstract.pt. (8411318) |
| 42 | 40 not 41 (3032) |
| 43 | (exp animal/ or nonhuman/) not exp human/ (9612770) |
| 44 | 42 not 43 (2919) |
| 45 | limit 44 to english language [Limit not valid in CDSR,DARE; records were retained] (2804) |
| 46 | 45 use emez (1249) |
| 47 | major depression/ (141961) |
| 48 | ((depression* or depressive* or melancholia* or paraphrenia* or psychos?s) adj2 (major or disorder* or involution* or unipolar*)).tw. (187247) |
| 49 | affective disorders/ (56371) |
| 50 | ((affective or mood) adj disorder*).tw. (90895) |
| 51 | anxiety disorders/ (48451) |
| 52 | bipolar disorder/ (97611) |
| 53 | ((anxiety adj (disorder* or neuros?s)) or ((bipolar or manic) adj (psychos?s or disorder* or depression* or depressive*))).tw. (165660) |
| 54 | or/47–53 (497656) |
| 55 | pharmacodynamics/ (22342) |
| 56 | (pharmacogenetic* or pharmacogenomic*).tw. (29773) |
| 57 | genetic testing/ (59139) |
| 58 | (((genetic* or gene or genes) adj2 (test or tests or testing or panel* or assess* or screen* or profil* or algorithm* or combinatorial)) or psychotropic panel*).tw. (204909) |
| 59 | (genotype adj (assignment* or method* or technique*)).tw. (948) |
| 60 | (genesight or assurex).tw. (25) |
| 61 | or/55–60 (292630) |
| 62 | 54 and 61 (3364) |
| 63 | case report/ or editorial.dt. or comment reply.dt. or letter.dt. (3980795) |
| 64 | 62 not 63 (3245) |
| 65 | limit 64 to english language [Limit not valid in CDSR,DARE; records were retained] (3106) |
| 66 | 65 use psyb (604) |
| 67 | 23 or 46 or 66 (2827) |
| 68 | 67 use pmoz (884) |
| 69 | 67 use emez (1249) |
| 70 | 67 use psyb (604) |
| 71 | 67 use coch (10) |
| 72 | 67 use cctr (71) |
| 73 | 67 use clhta (0) |
| 74 | 67 use cleed (3) |
| 75 | 67 use dare (6) |
| 76 | remove duplicates from 67 (1890) |
Appendix 3: Evidence Quality Assessment
Our first consideration was study design; we started with the assumption that randomized controlled trials are high quality, whereas observational studies are low quality. We then took into account five additional factors—risk of bias, inconsistency, indirectness, imprecision, and publication bias. Limitations in these areas resulted in downgrading the quality of evidence. Finally, we considered three main factors that may raise the quality of evidence: the large magnitude of effect, the dose-response gradient, and any residual confounding factors.26 For more detailed information, please refer to the latest series of GRADE articles.26
As stated by the GRADE Working Group, the final quality score can be interpreted using the following definitions:
| High | We are very confident that the true prognosis (probability of future events) lies close to that of the estimate |
| Moderate | We are moderately confident that the true prognosis (probability of future events) is likely to be close to the estimate, but there is a possibility that it is substantially different |
| Low | Our confidence in the estimate is limited: the true prognosis (probability of future events) may be substantially different from the estimate |
| Very Low | We have very little confidence in the estimate: the true prognosis (probability of future events) is likely to be substantially different from the estimate |
Table A3:
GRADE Evidence Profile for Comparison of GeneSight-Guided Care and Usual Care
| Number of Studies (Design) | Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Upgrade Considerations | Quality |
|---|---|---|---|---|---|---|---|
| Remission of Depression Symptoms | |||||||
| HAMD-17 | |||||||
| 1 (RCT) 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| QIDS-C16 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| PHQ-9 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| Response to Depression Therapy | |||||||
| HAMD-17 | |||||||
| 1 (RCT) 2 (observational) | Serious limitationsa | No limitations | No limitations | No limitations | None detected | None | ⊕⊕ Low |
| QIDS-C16 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| PHQ-9 | |||||||
| 1 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| Depression Score | |||||||
| HAMD-17 | |||||||
| 1 (RCT) 2 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| QIDS-C16 | |||||||
| 2 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| PHQ-9 | |||||||
| 2 (observational) | Serious limitationsa | No limitations | No limitations | Serious limitationsb | None detected | None | ⊕ Very low |
| Impact on Therapeutic Decisions | |||||||
| 1 (RCT) 3 (observational) | Serious limitationsa | Serious limitationsc | Very serious limitationsd | No limitations | None detected | None | ⊕ Very low |
| Patient Satisfaction | |||||||
| 1 (observational) | Serious limitationsa | No serious limitations | Very serious limitationse | No serious limitations | None detected | None | ⊕ Very low |
| Clinician Satisfaction | |||||||
| 1 (observational) | Serious limitationsa | No serious limitations | No serious limitations | No serious limitations | None detected | None | ⊕⊕ Low |
Abbreviations: GRADE, Grading of Recommendations Assessment, Development, and Evaluation; HAMD-17, 17-item Hamilton Rating Scale for Depression; PHQ-9, 9-item Patient Health Questionnaire; QIDS C16, 16-item Quick Inventory of Depressive Symptomatology, clinician rating; RCT, randomized controlled trial.
Risk of bias assessment details are available in Tables A2 and A3.
Wide confidence intervals impacted our confidence in the estimate of effect; experts advised that a clinically meaningful difference in HAMD-17 was two to three points.
Results were inconsistent between the studies: some demonstrated an increase in the mean number of medications per patient by end of study, and others a decrease.
Outcomes were indirect measures of the outcome of interest (the impact of guided therapy on therapeutic decision-making).
Measure of patient satisfaction was based on clinician perception of their patients' satisfaction.
Table A4:
Risk of Bias Among Randomized Controlled Trials Evaluating Pharmacogenomic Testing With GeneSight
| Author, Year | Allocation Concealment | Blinding | Complete Accounting of Patients and Outcome Events | Selective Reporting Bias | Other Limitations |
|---|---|---|---|---|---|
| Winner et al, 201329 | No limitationsa | Limitationsb | No limitationsc | None detected | Limitationsd |
Specific method of allocation was not reported, but study reported randomized allocation.
Patients and assessors were blinded to treatment allocation, but clinicians could not be blinded to whether or not they received the GeneSight Psychotropic test results and may have introduced bias during their interaction with patients. There was potential uncertainty around subjective outcomes such as patient-reported feelings of depression.
One patient in each study arm was lost to follow-up.
Study included authors with conflicts of interest related to Assurex, the manufacturer of the GeneSight Psychotropic test.
Table A5:
Risk of Bias Among Observational Studies Evaluating Pharmacogenomic Testing With GeneSight
| Author, Year | Appropriate Eligibility Criteria | Appropriate Measurement of Exposure | Appropriate Measurement of Outcome | Adequate Control for Confounding | Complete Follow-Up | Other Limitations |
|---|---|---|---|---|---|---|
| Hall-Flavin et al, 201228 | No limitationsa | No limitations | Limitationsb | Limitationsc | No limitationsd | Limitationse |
| Hall-Flavin et al, 201327 | No limitationsa | No limitations | Limitationsb | No limitationsf | Serious limitationsg | Limitationse |
| Winner et al, 201530 | No limitations | No limitations | Limitationsb | No limitationsh | Serious limitationsg | Limitationse |
Study included only patients with certain psychiatric disorders. This may have limited the generalizability of the results to other populations.
Outcomes that were subjective in nature, such as measurements of feelings of depression, could be at risk for sources of bias due to lack of blinding of patients and clinicians.
Baseline criteria were significantly different for patients in the guided and unguided groups for number of medications previously tried. Patients in the guided group had more attempts with different medications than those in the unguided group, and results were not adjusted for potential sources of confounding.
Only 72% of enrolled patients completed the study, but the authors conducted sensitivity analyses accounting for participant attrition using expectation maximization and last observation carried forward algorithms.
Study included authors with conflicts of interest related to Assurex, the manufacturer of the GeneSight Psychotropic test.
Study demonstrated balance in clinically meaningful criteria such as baseline depression score between treatment and control study arms.
Patients were dropped out after enrolment if they were prescribed a medication other than one covered by the GeneSight test, introducing a bias in effectiveness for the disease population as a whole.
Study conducted 5:1 propensity matching of unguided to guided patients and demonstrated no statistically significant difference in clinically meaningful baseline characteristics between the groups.
Author contributions
This report was developed by a multidisciplinary team from Health Quality Ontario. The lead clinical epidemiologist was Stacey Brener, the medical librarian was Corinne Holubowich.
KEY MESSAGES
Many people in Ontario live with mental illness, including major depressive disorder, bipolar disorder, schizophrenia, or anxiety. Medications are commonly used to treat mental illnesses, but choosing the right medication for each patient is a challenge. The GeneSight Psychotropic test uses a patient's unique genetic profile to guide doctors in choosing medications.
Patients with depressive symptoms who had the GeneSight test to help guide the choice of their medication responded better to treatment and their mood improved compared with patients who did not have the test. However, GeneSight-guided care did not lead to better rates of complete relief of depressive symptoms. Overall, these findings were based on evidence of low to very low quality and because of this, the effect of the GeneSight test on patient outcomes is uncertain.
Contributor Information
Health Quality Ontario:
About Health Quality Ontario
Health Quality Ontario is the provincial advisor on the quality of health care. We are motivated by a single-minded purpose: Better health for all Ontarians.
Who We Are
We are a scientifically rigorous group with diverse areas of expertise. We strive for complete objectivity, and look at things from a vantage point that allows us to see the forest and the trees. We work in partnership with health care providers and organizations across the system, and engage with patients themselves, to help initiate substantial and sustainable change to the province's complex health system.
What We Do
We define the meaning of quality as it pertains to health care, and provide strategic advice so all the parts of the system can improve. We also analyze virtually all aspects of Ontario's health care. This includes looking at the overall health of Ontarians, how well different areas of the system are working together, and most importantly, patient experience. We then produce comprehensive, objective reports based on data, facts and the voice of patients, caregivers and those who work each day in the health system. As well, we make recommendations on how to improve care using the best evidence. Finally, we support large scale quality improvements by working with our partners to facilitate ways for health care providers to learn from each other and share innovative approaches.
Why It Matters
We recognize that, as a system, we have much to be proud of, but also that it often falls short of being the best it can be. Plus certain vulnerable segments of the population are not receiving acceptable levels of attention. Our intent at Health Quality Ontario is to continuously improve the quality of health care in this province regardless of who you are or where you live. We are driven by the desire to make the system better, and by the inarguable fact that better has no limit.
REFERENCES
- (1).Patten S, Kennedy SH, Lam RW, O'Donovan C, Filteau M, Parikh SV, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults I. classification, burden and principles of management. J Affect Disord. 2009;117 suppl: S5–S14. [DOI] [PubMed] [Google Scholar]
- (2).Brien S, Grenier L, Kapral M.E., Kurdyak P, Vigod S. Taking Stock: A report on the quality of mental health and addictions services in Ontario. An HQO/ICES Report. Toronto, Health Quality Ontario and Institute for Clinical Evaluative Sciences; 2015. [Google Scholar]
- (3).Bouchard S, Verrier P. Anxiety disorders and comorbidities. Vancouver (BC): Anxiety Disorders Association of Canada; 2005. [Google Scholar]
- (4).Parikh SV, Segal Z, Grigoriadis S, Ravindran AV, Kennedy SH, Lam RW, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults II. Psychotherapy alone or in combination with antidepressant medication. J Affect Disord. 2009;117 suppl: S15–S25. [DOI] [PubMed] [Google Scholar]
- (5).Rotermann M, Sanmartin C, Hennessy D, Arthur M. Prescription medication use by Canadians aged 6 to 79. Health Reports. Catalogue no. 82-003-X ed. Ottawa (ON): Statistics Canada; 2014. p. 1–9. [PubMed] [Google Scholar]
- (6).Lam RW, Kennedy SH, Grigoriadis S, McIntyre R, Milev R, Ramasubbu R, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults III. Pharmacotherapy. J Affect Disord. 2009;117 suppl: S26–S43. [DOI] [PubMed] [Google Scholar]
- (7).Rush AJ, Trivedi MH, Wisniewski SR, Stewart JW, Nierenberg AA, Thase ME, et al. Bupropion-SR, sertraline, or venlafaxine-XR after failure of SSRIs for depression. N Engl J Med. 2006;354 (12): 1231–42. [DOI] [PubMed] [Google Scholar]
- (8).Anderson I, Haddad P. Canadian Network for Mood and Anxiety Treatments (CANMAT) clinical guidelines for the management of major depressive disorder in adults. Commentary. J Affect Disord. 2009;117 suppl: S3–S4. [DOI] [PubMed] [Google Scholar]
- (9).Fabbri C, Serretti A. Pharmacogenetics of major depressive disorder: top genes and pathways toward clinical applications. Curr Psychiatry Rep. 2015;17 (7): 50. [DOI] [PubMed] [Google Scholar]
- (10).Tansey KE, Guipponi M, Hu X, Domenici E, Lewis G, Malafosse A, et al. Contribution of common genetic variants to antidepressant response. Biol Psychiatry. 2013;73 (7): 679–82. [DOI] [PubMed] [Google Scholar]
- (11).Adkins DE, Souza RP, Aberg K, Clark SL, McClay JL, Sullivan PF, et al. Genotype-based ancestral background consistently predicts efficacy and side effects across treatments in CATIE and STAR*D. PLoS One. 2013;8 (2): e55239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chen PY, Wang SC, Poland RE, Lin KM. Biological variations in depression and anxiety between East and West. CNS Neurosci Ther. 2009;15 (3): 283–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Winner J, Allen JD, Altar CA, Spahic-Mihajlovic A. Psychiatric pharmacogenomics predicts health resource utilization of outpatients with anxiety and depression. Transl Psychiatry. 2013;3: e242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Drozda K, Muller DJ, Bishop JR. Pharmacogenomic testing for neuropsychiatric drugs: current status of drug labeling, guidelines for using genetic information, and test options. Pharmacotherapy. 2014;34 (2): 166–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Niitsu T, Fabbri C, Bentini F, Serretti A. Pharmacogenetics in major depression: a comprehensive meta-analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2013;45: 183–94. [DOI] [PubMed] [Google Scholar]
- (16).Altar CA, Carhart JM, Allen JD, Hall-Flavin DK, Dechairo BM, Winner JG. Clinical validity: Combinatorial pharmacogenomics predicts antidepressant responses and healthcare utilizations better than single gene phenotypes. Pharmacogenomics J. 2015;15 (5): 443–51. [DOI] [PubMed] [Google Scholar]
- (17).Murphy E, McMahon FJ. Pharmacogenetics of antidepressants, mood stabilizers, and antipsychotics in diverse human populations. Discov Med. 2013;16 (87): 113–22. [PMC free article] [PubMed] [Google Scholar]
- (18).Pitychoutis PM, Kokras N, Sanoudou D, Dalla C, Papadopoulou-Daifoti Z. Pharmacogenetic considerations for late life depression therapy. Expert Opin Drug Metab Toxicol. 2013;9 (8): 989–99. [DOI] [PubMed] [Google Scholar]
- (19).Bagdy G, Juhasz G, Gonda X. A new clinical evidence-based gene-environment interaction model of depression. Neuropsychopharmacol. 2012;14 (4): 213–20. [PubMed] [Google Scholar]
- (20).GeneSight Psychotropic [Internet]. Mason (OH): AssureRx Health Inc.; 2006. [updated 2016; cited 2016 Jan 18]. Available from: https://genesight.com/clinicians/genesight-tests/psychotropic/. [Google Scholar]
- (21).Genetic Testing Registry: GeneSight Psychotropic [Internet]. Bethesda, (MD): National Center for Biotechnology Information, US National Library of Medicine; 2014. [cited 2016 Jan 18]. Available from: http://www.ncbi.nlm.nih.gov/gtr/tests/508961/. [Google Scholar]
- (22).Local Coverage Determination: MoIDX: GeneSight Assay for refractory depression (L36325) [Internet]. Baltimore, (MD): Centers Medicare Services [updated 2015 Aug; cited 2016 Jun 1]. Available from: https://www.cms.gov/medicare-coverage-database/details/lcd-details.aspx?LCDId=36325&ContrId=358&ver=6&ContrVer=1&CntrctrSelected=358*1&Cntrctr=358&name=Noridian+Healthcare+Solutions%2c+LLC+%28Noridian+Healthcare+Solutions%2c+LLC+%2802402%2c+A+and+B+MAC%2c+J+-+F%29%29&LCntrctr=358*1&DocType=Active&bc=AAAAAAIAAAAAAA%3d%3d&.
- (23).Hicks JK, Bishop JR, Sangkuhl K, Muller DJ, Ji Y, Leckband SG, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98 (2): 127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Review Manager (RevMan) [computer program]. Version 5.3. Copenhagen: The Nordic Cochrane Centre, The Cochrane Collaboration;. 2008.
- (25).Crawford AA, Lewis G, Lewis SJ, Munafò MR. Systematic review and meta-analysis of serotonin transporter genotype and discontinuation from antidepressant treatment. Eur Neuropsychopharmacol. 2013;23: 1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Guyatt GH, Oxman AD, Schünemann HJ, Tugwell P, Knottnerus A. GRADE guidelines: a new series of articles in the Journal of Clinical Epidemiology. J Clin Epidemiol. 2011. Apr; 64 (4): 380–2. [DOI] [PubMed] [Google Scholar]
- (27).Hall-Flavin DK, Winner JG, Allen JD, Carhart JM, Proctor B, Snyder KA, et al. Utility of integrated pharmacogenomic testing to support the treatment of major depressive disorder in a psychiatric outpatient setting. Pharmacogenet Genomics. 2013;23 (10): 535–48. [DOI] [PubMed] [Google Scholar]
- (28).Hall-Flavin DK, Winner JG, Allen JD, Jordan JJ, Nesheim RS, Snyder KA, et al. Using a pharmacogenomic algorithm to guide the treatment of depression. Transl Psychiatry. 2012;2: e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Winner JG, Carhart JM, Altar CA, Allen JD, Dechairo BM. A prospective, randomized, double-blind study assessing the clinical impact of integrated pharmacogenomic testing for major depressive disorder. Discov Med. 2013;16 (89): 219–27. [PubMed] [Google Scholar]
- (30).Winner JG, Carhart JM, Altar CA, Goldfarb S, Allen JD, Lavezzari G, et al. Combinatorial pharmacogenomic guidance for psychiatric medications reduces overall pharmacy costs in a 1 year prospective evaluation. Curr Med Res Opin. 2015;31 (9): 1633–43. [DOI] [PubMed] [Google Scholar]
- (31).Hornberger J, Li Q, Quinn B. Cost-effectiveness of combinatorial pharmacogenomic testing for treatment-resistant major depressive disorder patients. Am J Manag Care. 2015;21 (6): e357–65. [PubMed] [Google Scholar]
- (32).Moher D, Liberati A, Tetzlaff J, Altman DG, the PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6 (6): e1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Assurex Health releases major update to Genesight Psychotropic test panel. Mason (OH): Assurex Health; 2013 [cited 2016 May 5]. Available from: https://assurexhealth.com/assurex-health-releases-major-update-to-genesight-psychotropic-test-panel/.
- (34).Zimmerman M, Martinez JH, Young D, Chelminski I, Dalrymple K. Severity classification on the Hamilton Depression Rating Scale. J Affect Disord. 2013;150 (2): 384–8. [DOI] [PubMed] [Google Scholar]
- (35).Reilly TJ, MacGillivray SA, Reid IC, Cameron IM. Psychometric properties of the 16-item Quick Inventory of Depressive Symptomatology: a systematic review and meta-analysis. J Psychiatr Res. 2015;60: 132–40. [DOI] [PubMed] [Google Scholar]
- (36).Volker D, Zijlstra-Vlasveld MC, Brouwers EP, Homans WA, Emons WH, van der Feltz-Cornelis CM. Validation of the Patient Health Questionnaire-9 for major depressive disorder in the occupational health setting. J Occup Rehabil. 2015;26 (2): 237–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Altar CA, Carhart J, Allen JD, Hall-Flavin D, Winner J, Dechairo B. Clinical Utility of Combinatorial Pharmacogenomics-Guided Antidepressant Therapy: Evidence from Three Clinical Studies. Mol Neuropsychiatry. 2015;1 (3): 145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Singh AB. Improved antidepressant remission in major depression via a pharmacokinetic pathway. Polygene Pharmacogenetic Report. 2015;13 (2): 150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]