Figure 6. Regulation of PP1-87B activity during mitosis.
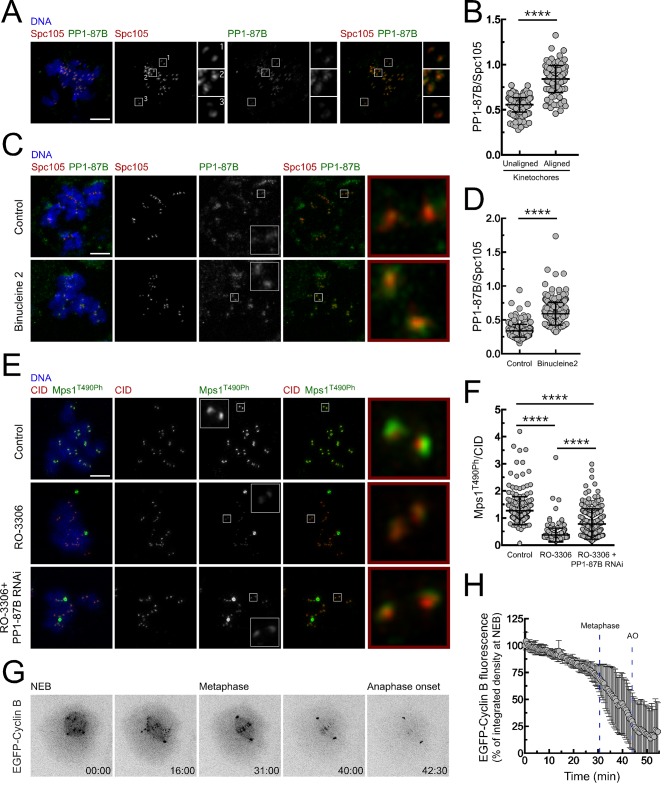
(A,B) Representative immunofluorescence images (A) and corresponding quantifications (B) of PP1-87B levels at kinetochores of unaligned and aligned chromosomes in S2 cells. PP1-87B fluorescence intensities were determined relative to Spc105 signal (N = 241 unaligned kinetochores and N = 110 aligned kinetochores from at least 6 cells). Scale bar: 5 μm. (C,D) Representative immunofluorescence images (C) and corresponding quantifications (D) of PP1-87B levels at unattached kinetochores of control and Binucleine 2-treated S2 cells. Cells were treated with MG132 (20 µM) for 1 hr followed by a 2 hr incubation with colchicine (30 µM) to generate unattached kinetochores. Binucleine 2 (20 µM) was added to cultures 30 min prior to colchicine treatment. PP1-87B fluorescence intensities were determined relative to Spc105 signal (N ≥ 282 kinetochores from at least 12 cells for each condition). Scale bar: 5 μm. (E,F) Representative immunofluorescence images (E) and corresponding quantifications (F) of Mps1T490Ph levels at unattached kinetochores of control and PP1-87B depleted cells in the presence of CDK1 inhibitor (RO-3306, 10 µM) for 1 hr. To generate unattached kinetochores cultured cells were incubated with colchicine (30 µM) for 2 hr. The insets display magnifications of the outlined regions. Mps1T490Ph fluorescence intensities were determined relative to CID signal (N ≥ 236 kinetochores from at least 10 cells for each condition). Scale bar: 5 μm. (G,H) Representative mitotic progression (G) and Cyclin B degradation profile (H) of S2 cells during an unperturbed mitosis. Mitotic progression of S2 cells expressing EGFP-Cyclin B was monitored by time-lapse microscopy and the integrated intensity of EGFP-Cyclin B fluorescence measured in all the frames. The values for EGFP-Cyclin B fluorescence at nuclear envelope breakdown (NEB) (time 0 min) were set to 100%. Dashed blue lines indicate the mean time from NEB to metaphase and to anaphase onset (AO) (N = 5 cells from a single experiment). Data information: in (B), (D), (F) and (H) data are presented as mean ± SD. Asterisks indicate that differences between mean ranks are statistically significant, ****p<0.0001 (Mann-Whitney U test). Numerical source data for this figure are provided in Figure 6—source data 1.
DOI: http://dx.doi.org/10.7554/eLife.25366.035