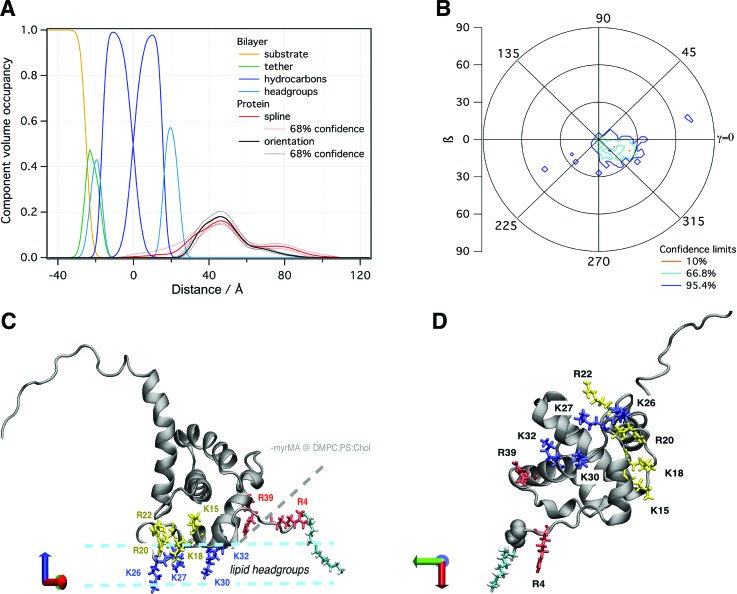
Fig. 5.
(a) NR CVO profile of myrMA on a 50:50 DOPS/DOPC stBLM at pH 8, 50 mM NaCl after incubation with 10 μM protein. The median protein envelope is shown with 68% confidence intervals (red traces). The median orientation fit using the MA NMR structure (PDB entry 2H3F) is shown for comparison (black trace). (b) Probability distribution of myrMA orientations with respect to the 50:50 DOPS/DOPC bilayer normal. (c) The most likely orientation of the protein (β ∼ 20°, γ ∼ 335°) on the membrane. Lysine residues (K26, K27, K30, and K32) that penetrate deeply into the lipid headgroup region are highlighted in blue. Basic residues with a peripheral interaction (K15 and R22) or slight penetration (K18 and R20) are shown in yellow. Arginine residues (R4 and R39) that were previously shown in (−myr)MA to interact closely with the membrane but are more peripheral in myrMA are shown in red. A myristate group, shown in cyan, was added to the protein structure to highlight its location. (d) Bottom view of myrMA in its membrane bound orientation.