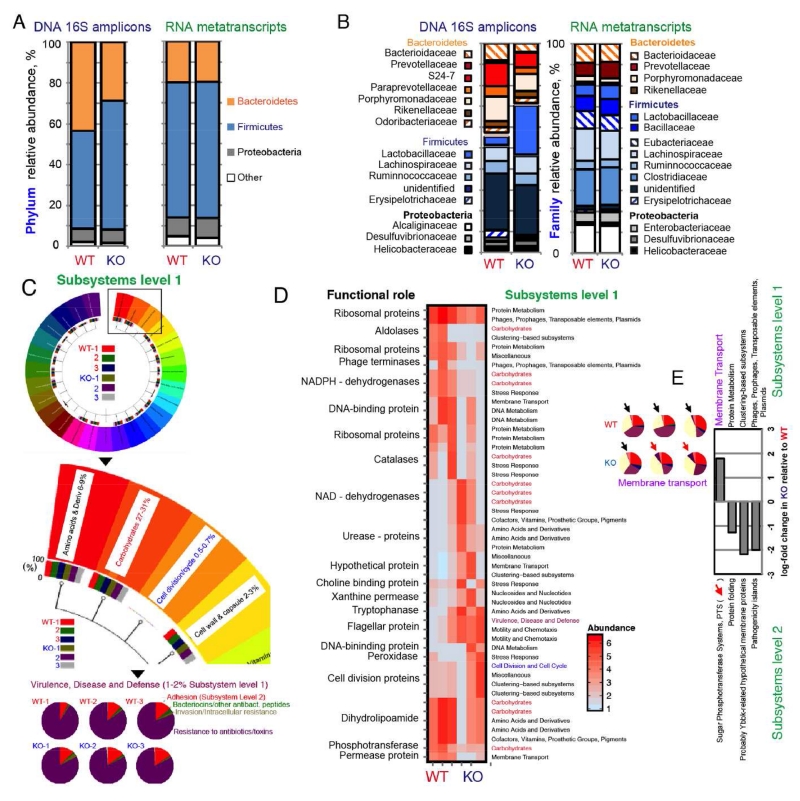
Figure 5. Gut metatranscriptome analysis indicates the Nod2 deletion did not have a significant impact on fecal microbiota composition and function.
Analysis on samples collected 7 days following IsPreFeH (3 mice/group). (A) Phylum-level composition based on 16S rRNA amplicon libraries and phylogenomic taxonomic inference from functional mRNA metatranscriptome data. (B) Family-level composition based on 16S rRNA amplicon libraries and phylogenomic taxonomic inference from functional mRNA metatranscriptome data (C) MG-RAST visualization of normalized transcript functional abundance (%) from the 6 individual metatranscriptomes. (D) Metatranscriptome cluster analysis of most expressed roles (specific enzymatic reactions). (E) Log-fold changes for the only 4 significantly detected functional roles (4 out of 193 level 2 categories). Pie charts, relative metatranscript abundance for the sugar phosphotransferase system.