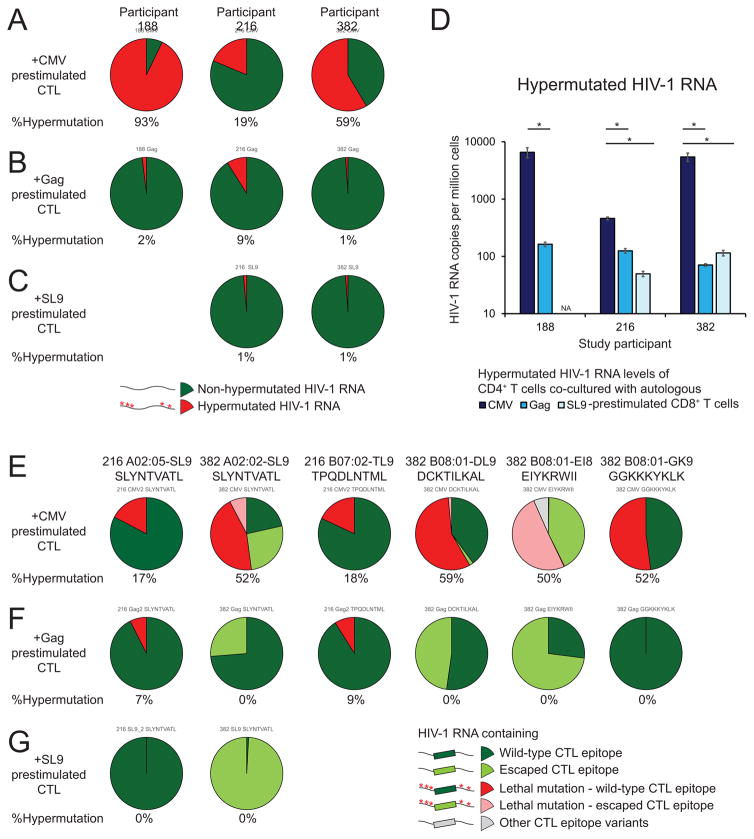
Figure 6. Cells containing HIV-1hypermut can be recognized by CD8+ T cells ex vivo.
Resting CD4+ T cells were activated with anti-CD3/CD28 in the presence of enfuvirtide and co-cultured with autologous CD8+ T cells prestimulated with a Gag peptide mixture, a CMV peptide mixture or a HLA-A2-restricted SL9 peptide. Targeted gag RNA deep sequencing and qRT-PCR was used to measure the proportion and the frequency of HIV-1 RNA containing lethal hypermutations. (A–D) Proportion (A–C) and frequency (D) of HIV-1 RNA containing lethal hypermutations from CD4+ T cells upon co-culture with autologous CMV- (A), Gag- (B), SL9-(C) prestimulated CD8+ T cells. (E–G) Proportion of HIV-1 RNA with or without lethal hypermutations 5′ to wild-type or escaped CTL epitopes upon co-culture with autologous CMV- (E), Gag- (F), and SL9- (G) prestimulated CD8+ T cells. Data represent mean ± SEM. *, P value <0.05 by two-tailed Student t-test.