Introduction
Foods and beverages produced by fermentation are essential to human nutrition worldwide and, therefore, have been extensively studied (Sõukand et al., 2015). Vinegar, kombucha beverage, milk kefir, water kefir, and cocoa are the products of acetic acid fermentation (Li et al., 2015). Acetic acid bacteria (AAB) oxidize sugars or ethanol to produce acetic acid, playing an important role in fermentation. AAB have been used historically for various fermentation processes and are Gram-negative obligate aerobic bacteria of the family Acetobacteraceae of Alphaproteobacteria (Saichana et al., 2015). Although various bacteria can produce acetic acid, most commercially used bacteria are species of Acetobacter, Gluconacetobacter, and Gluconobacter (Raspor and Goranovic, 2008). Among these organisms, Acetobacter species have attracted much attention in the field of biotechnology because these species are able to tolerate high acetic acid concentrations in the environment (Matsutani et al., 2011).
Acetobacter pasteurianus, one species of Acetobacter, has been used to brew vinegar worldwide (Gullo et al., 2006). The valuable and useful characteristics of A. pasteurianus motivated us to sequence and analyze the full genomes of two type strains of A. pasteurianus subspecies: A. pasteurianus subsp. ascendens LMG 1590T and A. pasteurianus subsp. paradoxus LMG 1591T. Type strain is usually the firstly isolated strain of the species, and exhibits all of the relevant phenotypic and genotypic properties cited in the species circumscriptions. Therefore, the genome sequence of type strain is important to analyze the phenotypic and genotypic characteristics of species (Kim et al., 2014). The genomes of these two strains were compared with other complete genome sequences, and the important proteins involved in acetic acid production are discussed.
Materials and Methods
Genomic DNA Isolation
Strains LMG 1590T and LMG 1591T, type strains of A. pasteurianus subsp. ascendens and A. pasteurianus subsp. paradoxus, respectively, were cultured for 3 days in YPGD media (0.5% yeast extract, 0.5% peptone, 0.5% glycerol, and 0.5% d-glucose) containing 4% (v/v) ethanol. Genomic DNA was extracted using phenol-chloroform extraction and ethanol precipitation. The quality of purified genomic DNA was tested by using NanoDrop 2000 UV–Vis spectrophotometer (Thermo Scientific, MA, USA) and Qubit 2.0 fluorometer (Life Technologies, MA, USA).
Genome Sequencing and Genome Comparison
The genomes of the two strains were sequenced at Macrogen using two different technologies: Illumina HiSeq and the PacBio single-molecule real-time technique with a 10-kb library (South Korea). De novo assembly of the read sequences was carried out using the hierarchical genome assembly process workflow. The annotation of the sequences was carried out using a modified version of the Prokka annotation pipeline, which incorporated Prodigal 2.60, Aragorn, and RNAmmer 1.2 for the prediction of open reading frames, tRNAs, and rRNAs, respectively (Seemann, 2014). Genome comparison among the two strains and other fully sequenced A. pasteurianus genomes was carried out by using Mauve software (Darling et al., 2010).
Direct Link to Deposited Data and Information to Users
The complete genome sequences have been deposited in GenBank under the accession numbers CP015164-CP015167 (A. pasteurianus subsp. ascendens LMG 1590T) and CP015168-CP015171 (A. pasteurianus subsp. paradoxus LMG 1591T) in October 2016. The BioProject designations for strains LMG 1590T and LMG 1591T are PRJNA3221271 and PRJNA317328,2 respectively. A. pasteurianus subsp. ascendens LMG 1590T and A. pasteurianus subsp. paradoxus LMG 1591T are available from the BCCM/LMG Bacteria Collection under accession numbers LMG 1590 and LMG 1591, respectively.
Interpretation of Data Set
General Genome Sequence Property
We obtained 218,360 raw reads covering a total of 1,387,777,653 bp with 228× genome coverage for strain LMG 1590T and 146,922 raw reads covering a total of 897,929,341 bp with 124× genome coverage for strain LMG 1591T. The complete genome sequence of strain LMG 1590T contained a circular chromosome of 2,859,878 bp with 53.1% G + C content and three circular plasmids with 55.4% G + C content. The genome sequence of LMG 1591T was also assembled into a circular chromosome of 2,810,721 bp with 53.2% G + C content and three circular plasmids with 54.3% G + C content. The general features of the genomes are summarized in Table 1. Briefly, the analyses of the strain LMG 1590T genome identified 2,931 genes. Among them 2,856 genes were annotated as coding DNA sequences (CDSs). A total of 3,163 genes were predicted from the genome of strain 1591T of which 3,088 genes were identified as CDSs. The genome sequences data are available in FASTA, annotated GenBank flat file, graphical, and ASN.1 formats.
Table 1.
Genome features of the Acetobacter pasteurianus subsp. strains LMG 1590T and LMG 1591T.
| Features | LMG 1590T | LMG 1591T |
|---|---|---|
| Chromosome | ||
| Contig number | 1 | 1 |
| Size (bp) | 2,859,878 | 2,810,721 |
| G + C (%) | 53.1 | 53.2 |
| Total genes | 2,784 | 2,760 |
| Coding DNA sequence (CDS) | 2,709 | 2,685 |
| tRNA | 56 | 56 |
| rRNA | 15 | 15 |
| Other RNA | 4 | 4 |
| Plasmid | ||
| Number | 3 | 3 |
| Size (bp) | 49,380/46,811/43,148 | 259,464/117,661/28,186 |
| G + C (%) | 55.4 | 54.3 |
| Total genes | 147 | 403 |
| CDS | 147 | 403 |
| tRNA | 0 | 0 |
| rRNA | 0 | 0 |
| Other RNA | 0 | 0 |
Genome Comparison
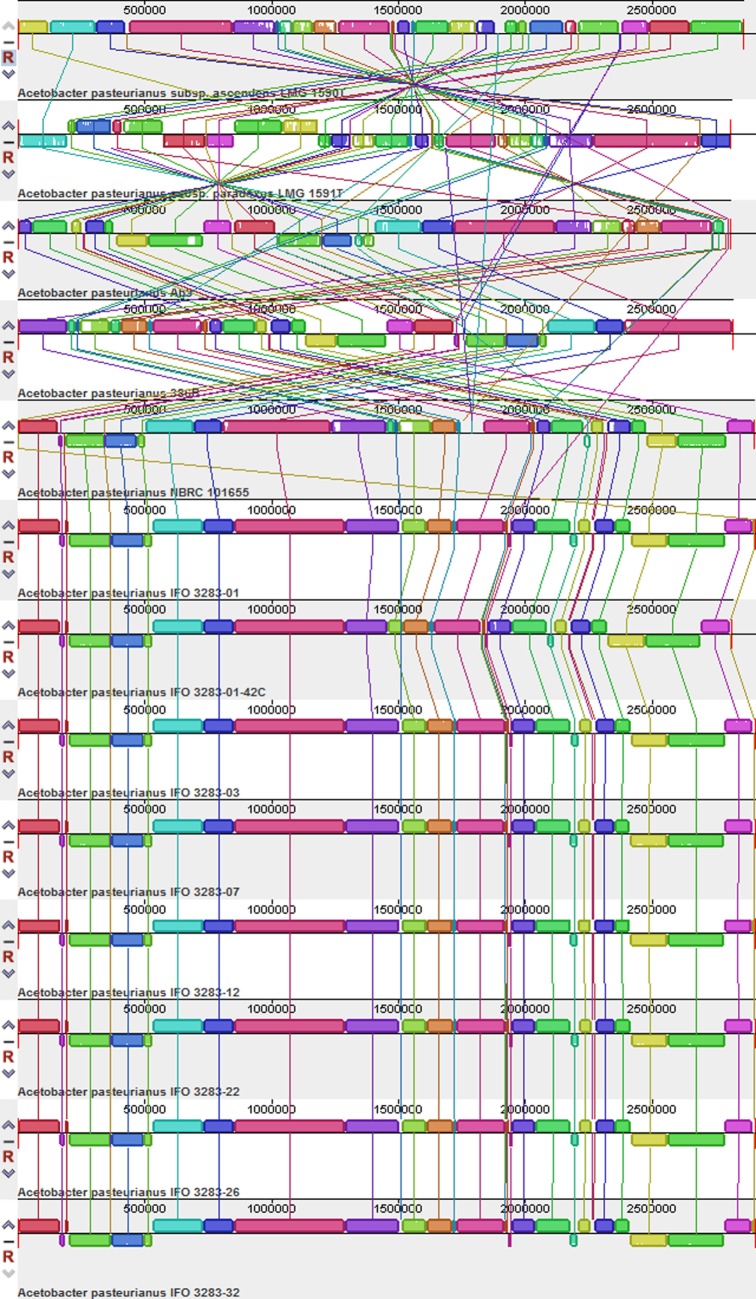
To investigate the overall genomic differences between the sequenced A. pasteurianus strains, including LMG 1590T and LMG 1591T, and the previously sequenced A. pasteurianus species, a global alignment of genome sequences from 13 strains was performed using Mauve software (Darling et al., 2010). The results showed that A. pasteurianus NBRC 101655, A. pasteurianus 386B, and eight A. pasteurianus IFO strains were quite similar with respect to genome structure in the chromosome (Figure 1), which was inconsistent with previous reports (Illeghems et al., 2013; Wang et al., 2015). Moreover, rearrangements, deletions, amplifications, and insertions occurred frequently in A. pasteurianus Ab3, A. pasteurianus subsp. ascendens LMG 1590T, and A. pasteurianus subsp. paradoxus LMG 1591T (Figure 1). Similar phenomena were also observed in the plasmids of these strains (Figure S1 in Supplementary Material).
Figure 1.
Global multiple alignments of Acetobacter pasteurianus chromosomes. The 13 genomes were compared to each other using progressive MAUVE with default parameters. Colored blocks outline the genome sequence that aligned to part of another genome and was presumably homologous and internally free from genomic rearrangement (locally collinear blocks). White regions are sequences that were not aligned and probably contained sequence elements specific to a particular genome. Blocks below the center line indicate regions that aligned in the reverse complement (inverse) orientation. The names of the strains are listed at the bottom of the blocks.
Acetic Acid-Producing Enzymes
The pyrroloquinoline quinone (PQQ)-alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH) are responsible for the oxidative metabolism of ethanol to produce acetic acid in AAB. These proteins generally consist of three subunits: a quinohemoprotein catalytic subunit, a triheme cytochrome c subunit, and a third subunit with unknown function. Sequence analysis of the catalytic subunits of the ADHs from strains LMG 1590T and LMG 1591T by a combined transmembrane topology and signal peptide predictor indicated that both of the proteins had a signal peptide and were located in the periplasmic space (Käll et al., 2004). The ADHs were aligned with the well-studied ADH from Pseudomonas putida (50% identity to ADHs from LMG 1590T and LMG 1591T) (Xia et al., 1996), and the final output was processed using the program ESPript 3.0 (Robert and Gouet, 2014). The results indicated that the PQQ bound to the N-terminal portion, whereas the C-terminal end bound the heme c (Figure S2 in Supplementary Material). The ADHs and ALDHs from the 13 strains were further aligned using Clustal Omega (Sievers and Higgins, 2014). Both ADHs and ALDHs from the strains showed high identity (>98%; Tables S1 and S2 in Supplementary Material), although Acetobacter species are known to exhibit genetic instability (Azuma et al., 2009). Because ethanol and acetic acid tolerance could be partly attributed to the intrinsic properties of the amino acid sequences of the two proteins and high concentrations of ethanol would not cause mutations in their sequences (Trcek et al., 2006; Zheng et al., 2015), we proposed that the high conservation of the proteins may contribute to the stable industrial performance of A. pasteurianus.
In conclusion, the complete genomes of two A. pasteurianus subspecies were sequenced and assembled into one chromosome and three plasmids. Comparative genome and sequence analyses showed that rearrangements occurred in the A. pasteurianus strains and that the ADHs and ALDHs responsible for acetic acid production were highly conserved in these strains.
Author Contributions
BC performed the experiments. BJ and CJ analyzed the data and wrote the manuscript. GC, KK, JM, and S-HY helped in data analysis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer, FF, and handling editor declared their shared affiliation, and the handling editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
This work was supported by the Cooperative Research Program for Agriculture Science and Technology Development (project nos. PJ00999302 and PJ01090604) Rural Development Administration, Republic of Korea.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fbioe.2017.00033/full/#supplementary-material.
References
- Azuma Y., Hosoyama A., Matsutani M., Furuya N., Horikawa H., Harada T., et al. (2009). Whole-genome analyses reveal genetic instability of Acetobacter pasteurianus. Nucleic Acids Res. 37, 5768–5783. 10.1093/nar/gkp612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling A. E., Mau B., Perna N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. 10.1371/journal.pone.0011147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullo M., Caggia C., De Vero L., Giudici P. (2006). Characterization of acetic acid bacteria in “traditional balsamic vinegar”. Int. J. Food Microbiol. 106, 209–212. 10.1016/j.ijfoodmicro.2005.06.024 [DOI] [PubMed] [Google Scholar]
- Illeghems K., De Vuyst L., Weckx S. (2013). Complete genome sequence and comparative analysis of Acetobacter pasteurianus 386B, a strain well-adapted to the cocoa bean fermentation ecosystem. BMC Genomics 14:526. 10.1186/1471-2164-14-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käll L., Krogh A., Sonnhammer E. L. L. (2004). A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338, 1027–1036. 10.1016/j.jmb.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Kim B. S., Yi H., Chun J., Cha C. J. (2014). Genome sequence of type strain of Staphylococcus aureus subsp. aureus. Gut Pathog. 6, 6–6. 10.1186/1757-4749-6-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Li P., Feng F., Luo L.-X. (2015). Microbial diversity and their roles in the vinegar fermentation process. Appl. Microbiol. Biotechnol. 99, 4997–5024. 10.1007/s00253-015-6659-1 [DOI] [PubMed] [Google Scholar]
- Matsutani M., Hirakawa H., Yakushi T., Matsushita K. (2011). Genome-wide phylogenetic analysis of Gluconobacter, Acetobacter, and Gluconacetobacter. FEMS Microbiol. Lett. 315, 122–128. 10.1111/j.1574-6968.2010.02180.x [DOI] [PubMed] [Google Scholar]
- Raspor P., Goranovic D. (2008). Biotechnological applications of acetic acid bacteria. Crit. Rev. Biotechnol. 28, 101–124. 10.1080/07388550802046749 [DOI] [PubMed] [Google Scholar]
- Robert X., Gouet P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saichana N., Matsushita K., Adachi O., Frébort I., Frebortova J. (2015). Acetic acid bacteria: a group of bacteria with versatile biotechnological applications. Biotechnol. Adv. 33, 1260–1271. 10.1016/j.biotechadv.2014.12.001 [DOI] [PubMed] [Google Scholar]
- Seemann T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- Sievers F., Higgins D. G. (2014). Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol. Biol. 1079, 105–116. 10.1007/978-1-62703-646-7_6 [DOI] [PubMed] [Google Scholar]
- Sõukand R., Pieroni A., Biró M., Dénes A., Dogan Y., Hajdari A., et al. (2015). An ethnobotanical perspective on traditional fermented plant foods and beverages in Eastern Europe. J. Ethnopharmacol. 170, 284–296. 10.1016/j.jep.2015.05.018 [DOI] [PubMed] [Google Scholar]
- Trcek J., Toyama H., Czuba J., Misiewicz A., Matsushita K. (2006). Correlation between acetic acid resistance and characteristics of PQQ-dependent ADH in acetic acid bacteria. Appl. Microbiol. Biotechnol. 70, 366–373. 10.1007/s00253-005-0073-z [DOI] [PubMed] [Google Scholar]
- Wang B., Shao Y., Chen T., Chen W., Chen F. (2015). Global insights into acetic acid resistance mechanisms and genetic stability of Acetobacter pasteurianus strains by comparative genomics. Sci. Rep. 5, 18330. 10.1038/srep18330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z.-X., Dai W.-W., Zhang Y.-F., White S. A., Boyd G. D., Mathews S. F. (1996). Determination of the gene sequence and the three-dimensional structure at 2.4 Å resolution of methanol dehydrogenase from MethylophilusW3A1. J. Mol. Biol. 259, 480–501. 10.1006/jmbi.1996.0334 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Zhang K., Su G., Han Q., Shen Y., Wang M. (2015). The evolutionary response of alcohol dehydrogenase and aldehyde dehydrogenases of Acetobacter pasteurianus CGMCC 3089 to ethanol adaptation. Food Sci. Biotechnol. 24, 133–140. 10.1007/s10068-015-0019-x [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.