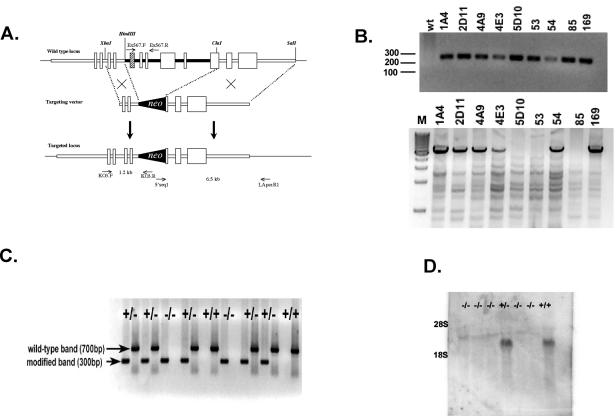
FIG. 1.
Targeted disruption of the Prss16 by homologous recombination. (A) A neomycin resistance cassette was inserted between the HindIII and ClaI sites replacing exon 5 (hatched box), which includes the putative active serine residue, through exon 9 and the 5′ end of exon 10. (B) 5′ (upper panel) and 3′ (lower panel) PCR analysis of ES cells. Primers KO3.F and KO3.R were used to screen for 5′ integration of the targeting vector. 3′ integration was confirmed by PCR with primers 5′seq1 and LApcr.R1. M, molecular weight markers (in hundreds); wt, wild type. (C) Genotype analysis of offspring from heterozygous parents of modified Prss16. Tail DNA was used for PCR analyses of the mutant and wild-type alleles with primer pairs KO3.F-KO3.R (lanes 1, 3, 5, 7, 9, 11, 13, 15, 17) and Ex567.F-Ex567.R (lanes 2, 4, 6, 8, 10, 12, 14, 16, 18), respectively. (D) Prss16−/− mice do not produce Prss16 mRNA. Northern blot of total thymus RNA from Prss16−/−, Prss16+/−, and Prss16+/+ mice probed with a Prss16 cDNA.