Fig. 1.
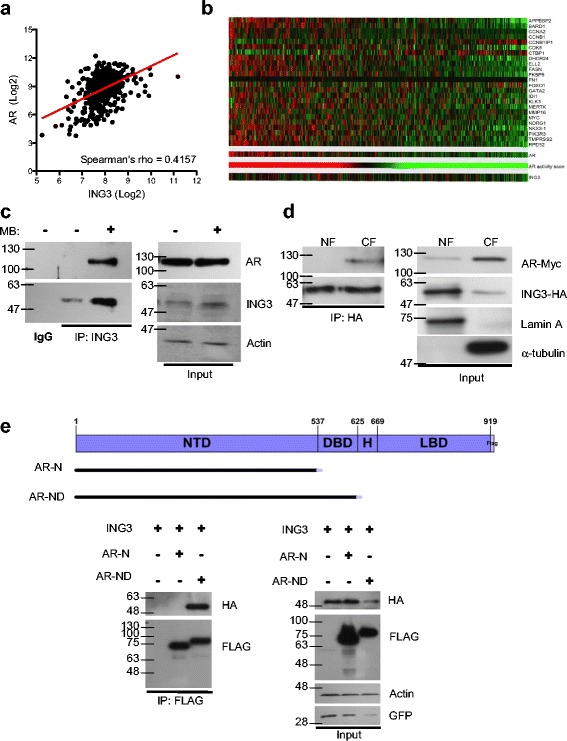
ING3 interacts with the AR. a The RNAseq data from TCGA prostate adenocarcinoma cohort (n = 498 cancer cases) was retrieved, levels of ING3 and AR mRNA in tumor samples were plotted in log2 scale, and linear regression was graphed. b Levels of ING3 and 25 androgen-regulated genes from TCGA RNAseq data were analyzed. AR activity score was generated by summation of z-scores, and patients were organized based on AR activity. c Endogenous AR-ING3 interactions were assessed by co-precipitation in LNCaP cells +/– 10 nM MB. d HEK293T cells were transfected with ING3-HA and AR plasmids in the absence of MB. Nuclear and cytosolic fractions were isolated according to our Rapid, Efficient, and Practical (REAP) protocol [34] and IPs were performed using HA-bound beads to pull down ING3 in each fraction. The IP samples were then subjected to western blotting by standard methods to detect co-precipitated proteins. Lamin A and α-tubulin were used as nuclear and cytosolic markers, respectively. e AR deletion constructs used to map domains required for interaction are shown. Co-precipitation study using HEK293T cells co-transfected with AR deletion mutants and ING3-HA. FLAG beads were used to precipitate AR constructs, and ING3 was detected in IP samples by western blotting
