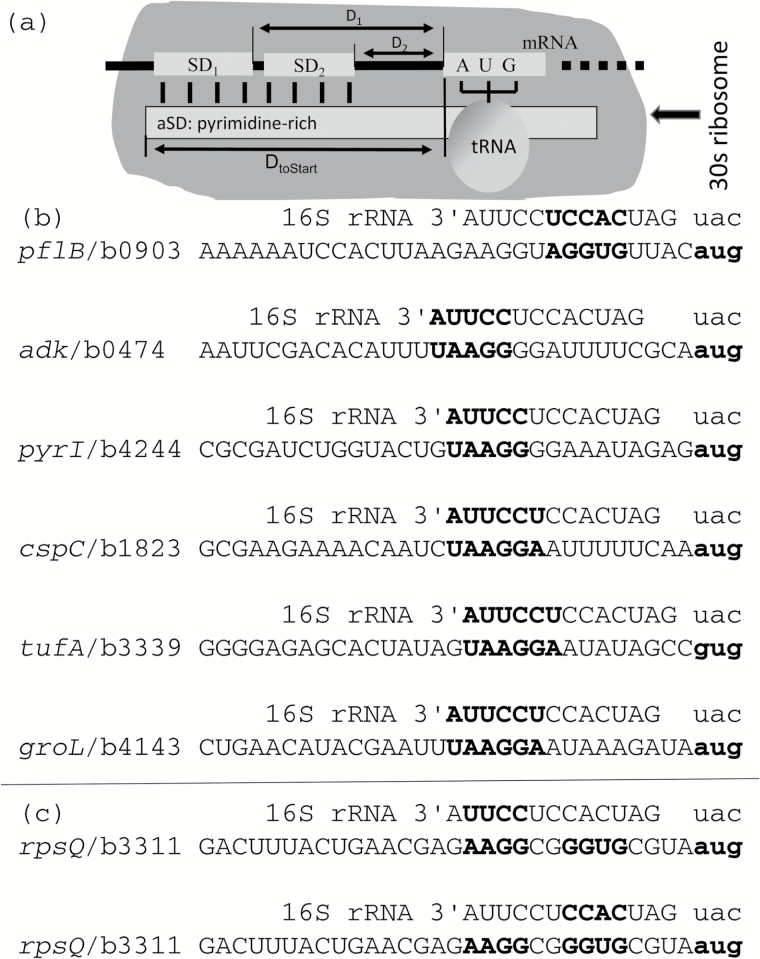
Figure 1.
Key features of interacting components for juxtaposing the start codon against the anticodon of the initiating fMet-tRNAfMet. (a) A model of SD/aSD pairing. Two different SDs on mRNA (SD1 and SD2), with different distances (D1 and D2, respectively) to the initiation codon AUG can both properly align AUG against the tRNA. The 2 different SD/aSD pairings result in the same DtoStart, defined as the distance between th’e 3′ end of ssu rRNA to the start codon. (b) A sample of SD/aSD pairing from highly expressed Escherichia coli genes, with the start codon and the tRNA anticodon in small case. (c) One example of a highly expressed gene (rpsQ) with 2 putative SDs (c). Gene IDs are in the form of “gene name/Locus_tag”. SDs in (b) and (c) differ in sequence and distance to the start codon, but they all have similar DtoStart. A change in DtoStart will lead to misalignment of start codon and tRNA anticodon.