Fig. 1.
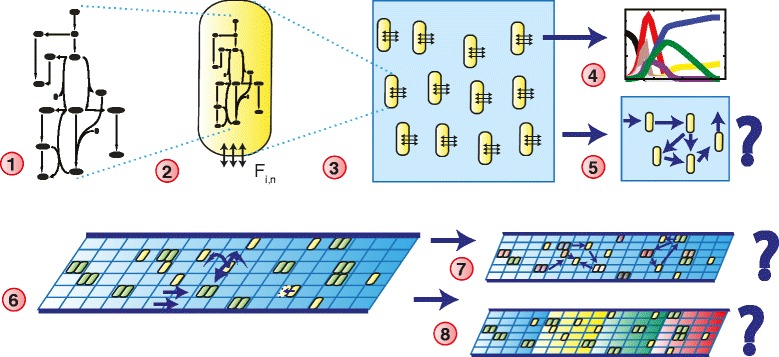
Workflow of the modeling. (1) Construction of “metabacterium” model, based on a Lactobacillus plantarum GEM [28] extended with metabolic pathways commonly found in the gut microbiota; (2) dynamic flux-balance analysis model; (3) well-mixed community of “metabacteria” exchanging metabolites via a common medium; (4) observation of metabolites in the common medium; (5) measure cross-feeding coefficient; (6) spatial modeling in a gut-like environment with evolving “metabacteria”; (7) look for speciation and cross-feeding; (8) look for stratification of metabolic environment
