Fig. 3.
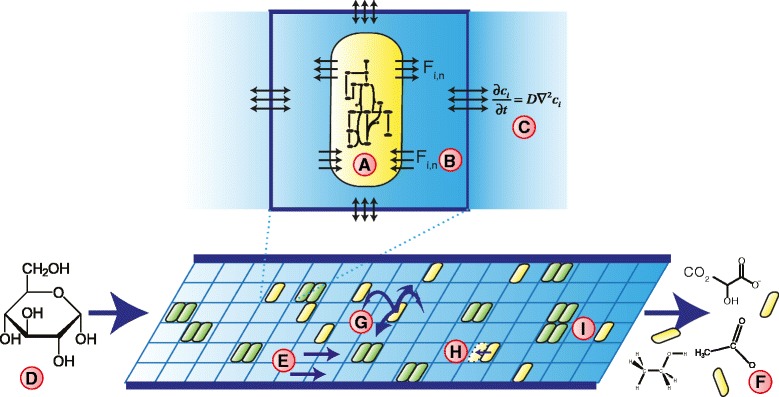
Setup of the simulation model of a metabolizing gut microbial community. The model represents a community of growing subpopulations of genetically identical bacteria. a The metabolism of each population is modeled using a unique, modified GEM of L. plantarum[28]; b Based on extracellular metabolite concentrations, the genome scale model predicts the growth rate (r) of the subpopulation and the influx and efflux rates of a subset of 115 metabolites. These are used as derivatives for a partial-differential equation model describing the concentrations of extracellular metabolites, , where c the metabolites diffuse between adjacent grid sites, . d The population is represented on a two-dimensional, tube-like structure, with periodic inputs of glucose. e To mimic advection of metabolites through the gut, the concentrations are periodically shifted to the right, until they f exit from the end of the tube. g The bacterial populations hop at random to adjacent grid sites; to mimic adherence to the gut wall mucus bacterial populations are not advected, unless indicated otherwise. h Once the subpopulation has grown to twice its original size, it divides into an empty spot in the same lattice size at which time the metabolic network is mutated. i Two subpopulations can live on one grid point; with yellow indicating presence of one subpopulation, and green indicating the presence of two subpopulations. (Structural formulas: Licensed under Public domain via Wikimedia Commons; “Alpha-D-Glucopyranose” by NEUROtiker, also licenced under public domain via Wikipedia Commons)
