Figure 1. Dissecting the combinatorial control logic underlying the pathogen-responsive transcriptome.
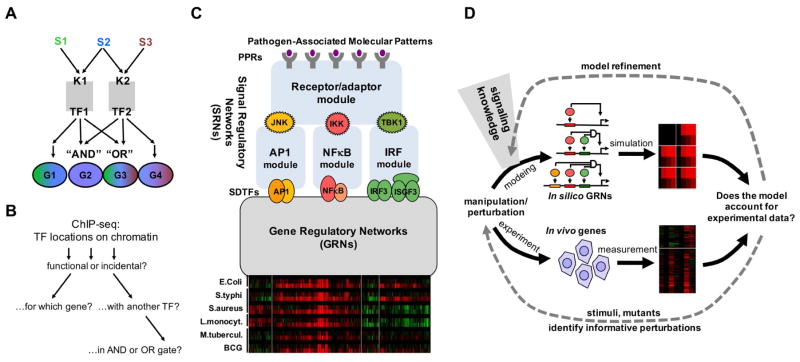
(A) Schematic of how combinatorial control of kinases (K) signal-dependent transcription factors (TFs) can mediate stimulus (S)-specific gene (G) expression programs. In this example the presence of AND and OR gates allows two TFs to mediate four distinct stimulus-responsive gene expression programs.
(B) While ChIP-seq studies enable genome-wide location analysis of TFs, it remains challenging to determine whether these binding events are functional or incidental, which gene or transcription start site they may regulate, and whether they function in conjunction with other TFs nearby or at a distance, in potential AND or OR gates.
(C) Schematic of key signal regulatory networks (SRNs) that control pathogen-responsive gene expression programs, exemplified by clusters of co-regulated genes (re-analysis of data by (Nau et al., 2002)). Prior studies of these SRNs have resulted in computational models of the molecular mechanisms (blue boxes) that recapitulate observed stimulus-responsive activation of the signal-dependent transcription factors (SDTFs) AP1, NFκB, and IRF. How these SDTF activities are interpreted to produce pathogen responsive gene expression programs is determined by the gene regulatory networks (GRNs), for which no mechanistic model has been developed (grey box).
(D) Schematic of the workflow for iteratively refining a mathematical model that recapitulates pathogen-responsive inflammatory and immune gene expression programs. Model formulations of increasing complexity are constructed and refined based on prior knowledge of the signaling network, and assessed by comparison to experimental data. With each round of simulation and experimentation of additional conditions the model is iteratively refined.