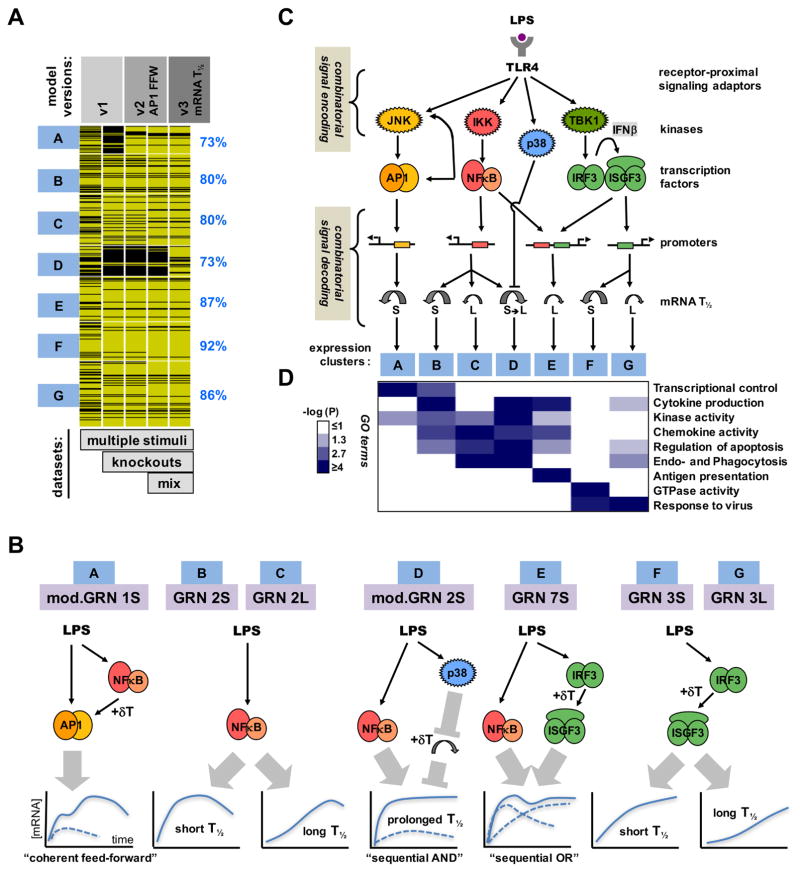
Figure 7. Iterative modeling of pathogen-responsive GRNs reveals combinatorial control mediated by sequentially acting regulatory mechanisms.
(A) Tracking the performance of iterative models with increasing datasets. Genes associated with expression clusters A to G passed (yellow lines) or did not pass (black lines) the Spearman goodness of fit criterion in successive GRN model versions and an increasing number of experimental “-omic” datasets generated in indicated perturbation studies, such as “multiple stimuli” (Figure 2), “knockouts” (Figure 3), and “mixed” stimuli (Figure 4). The fraction of genes that pass the goodness of fit Spearman correlation criterion for model v3 (including mod.GRN1S and mod.GRN2S) is shown on the right.
(B) Schematics of the GRNs that govern expression of genes in clusters A to G in MEFs and macrophages. (Schematic expression profiles are indicated as a solid blue line for wild-type cells, or as dashed lines in cells lacking one of the modifying pathway.) Cluster A expression is driven by AP1 which is controlled by a “coherent feed-forward” gate involving NFκB-driven autocrine PDGF which introduces a delay relative to the direct activation pathway. Clusters B, C and D are NFκB-driven, but specificity is generated by distinct mRNA half-life control. LPS-responsive p38-mediated inhibition of mRNA decay combines with LPS-responsive NFκB-mediated mRNA synthesis to form a “sequential AND” gate in the GRN for cluster D. Cluster E is driven by NFκB and IRF, forming a “sequential OR” gate between a rapidly activated immediate-early TF (NFκB) and a delayed, protein-synthesis-dependent TF (ISGF3). Cluster F and G are ISGF3-driven, but specificity is generated by distinct mRNA decay rates.
(C) A summary schematic of signal encoding and decoding via SRNs and GRNs, respectively. Combinatorially encoded signals are decoded not only by the promoter architecture of TF binding sites, but also by signal-responsive mRNA half-life control.
(D) Gene expression clusters are associated with distinct physiological functions. Highly enriched gene ontology (GO) terms were evaluated based on -log10 p-values as indicated on the white-blue scale.