Figure 3.
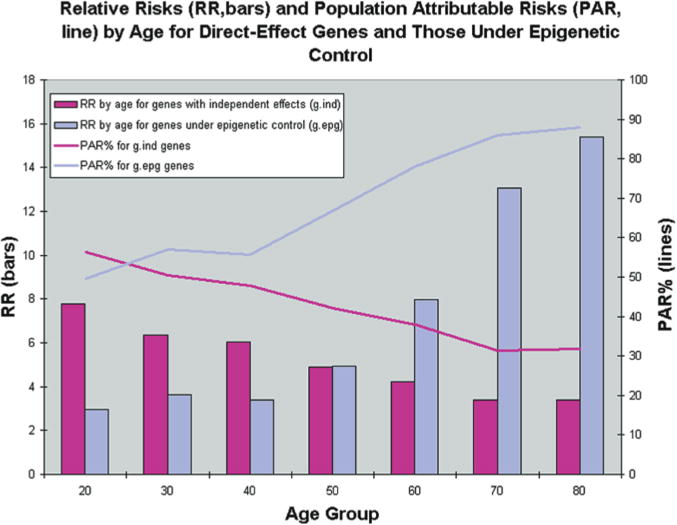
Results from simulations of 40 populations. Simulations were used to create 40 populations containing affected and unaffected individuals for a genetic epidemiologic analysis. Age, environmental status, and genotypes for three different genes were first simulated at random according to specified frequencies. Disease status was then simulated according to CDGE. From these simulated populations, relative risks for gind and gdep genes were estimated in cross-sectional analysis by each age decade, and averaged over 40 populations (bars). Because this risk reflects only the magnitude of a genetic effect, and not the importance of that genotype with respect to all cases in the population, we also estimated population attributable risk percentage (PAR%) at each decade. This reflects the proportion of cases in the population that can be explained by the particular genetic effect. (Reprinted, with permission, from Bjornsson et al. 2004 [©Elsevier].)
