Figure 5.
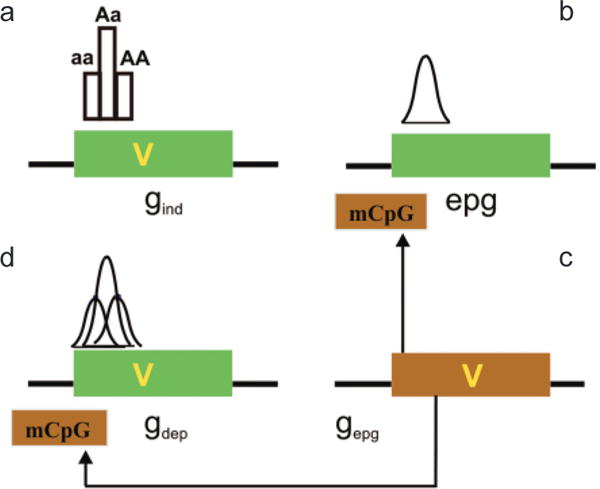
Interaction of genetic and epigenetic variation at a genomic level. (a) A gene (green box) can contain a sequence variant (V) that contributes to disease. Gene variants that are not influenced by epigenetic modification in their disease contribution (although epigenetics can contribute to normal function) are epigenetic independent (gind). The distribution of phenotype of a single locus will not be Gaussian in the absence of other factors, e.g., environmental. (b) Epigenetic variation can contribute to disease phenotype directly, independent of a genetic variation in the target gene (epg). Epigenetic variation itself is quantitative and thus can impart the quantitative nature to a trait, even at a single locus. (c) If the penetrance of a gene sequence variant is affected by epigenetic modification (mCpG), the gene is “epigenetic dependent” (gdep). In this case, the genetic and epigenetic variation together could contribute to a Gaussian distribution, even at a single locus. Note that the epigenetic modification is drawn on the gene but it could be at some distance upstream or downstream from that gene. The epigenetic modification need not be methylation, which is drawn here for convenience. (d) A genetic variant that can influence this epigenetic modification (e.g., encoding a chromatin-modifying protein) is referred to as gepg, and its influence is denoted by arrows. (Reprinted, with permission, from Bjornsson et al. 2004 [©Elsevier].)
