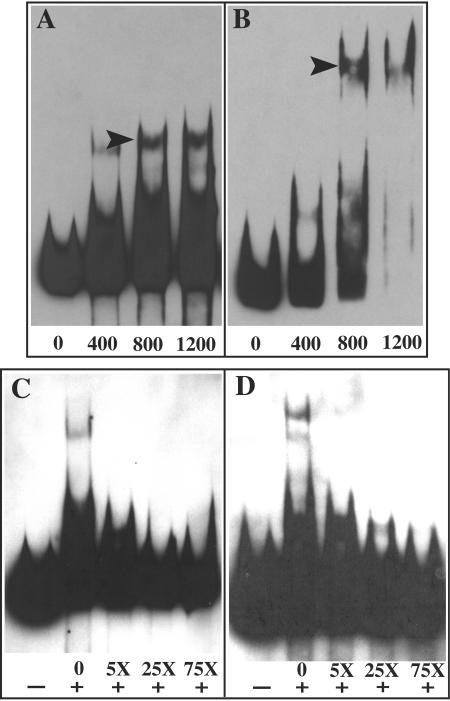
FIG. 4.
Binding specificity of chlamydial DcrA to selected DNA templates by EMSA. Increasing concentrations of purified DcrA (400, 800, and 1,200 nM) were added to 50 fmol of biotin-labeled plasmid inserts from pJER178 (A) and pJER406 (B) to illustrate a concentration-dependent increase in binding. Arrowheads indicate multimeric DcrA-DNA complexes. To examine competitive inhibition of chlamydial DcrA binding to selected DNA templates, 800 nM purified DcrA was absent (−) or present (+) in EMSA reaction buffer and 50 fmol of biotin-labeled substrate from pJER178 (C) and pJER406 (D) was incubated with a 5-, 25-, or 75-fold molar excess of unlabeled substrate. EMSA images were scanned into Adobe Photoshop 7.0 with a Microtek Scan Maker 5, embedded, assembled, and exported in Adobe Illustrator 10 as EPS and TIFF files.