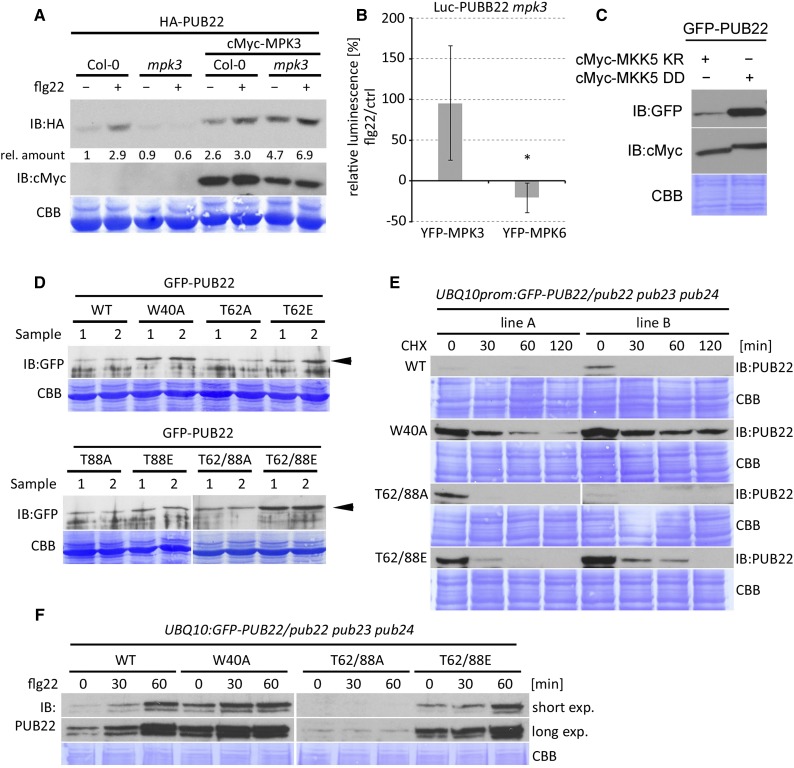
Figure 3.
PUB22 Stabilization Is Dependent on MPK3.
(A) HA-PUB22 was expressed alone or with cMyc-MPK3 in Col-0 and mpk3 background and treated with DMSO or flg22 (100 nM) for 45 min in Arabidopsis protoplasts. Band intensities were determined and amounts relative to HA-PUB22 in Col-0 calculated. The experiment was repeated with similar results.
(B) Luciferase (Luc) PUB22 fusion was coexpressed with MPK3 or MPK6 in mpk3 protoplasts. Luminescence was measured in control (water) and flg22 (100 nM) treated samples after 30 min. Relative luminescence to control samples after flg22 is shown as average ± sd obtained from three independent experiments. Statistical significance was assessed by Student’s t test, *P < 0.05.
(C) GFP-PUB22 was coexpressed with cMyc-MKK5 AspAsp (active) or cMyc-MKK5 LysArg (inactive) versions in Arabidopsis protoplasts. Samples were taken after 8 h incubation. The experiment was repeated three times with similar results.
(D) GFP-PUB22 wild-type and mutant variants were constitutively expressed in Arabidopsis protoplasts to test protein accumulation. Shown are samples from two independent transformations. Arrowhead highlights PUB22 and mutant variants.
(E) UBQ10:GFP-PUB22/pub22 pub23 pub24 independent transgenic lines (A and B) constitutively expressing GFP-PUB22 wild-type and mutant variants were treated with CHX (50 µM), and samples were taken at the indicated times. The experiment was repeated three times with similar results.
(F) UBQ10:GFP-PUB22/pub22 pub23 pub24 transgenic seedlings constitutively expressing GFP-PUB22 wild-type and variants were treated with flg22 (1 µM) for the indicated times to determine protein stabilization. The experiment was repeated three times with similar results.