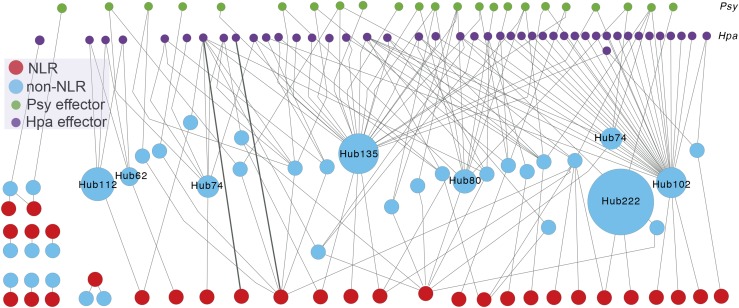
Figure 2.
The Interconnected Activity of NLRs and Protein Interaction Hubs.
A network illustrating interactions among pathogen effectors, effector targets, and NLRs reconstructed from the PPIN-1 interactome study by Mukhtar et al. (2011). The reconstruction using Cytoscape visualizes complexity of the interactions that NLR proteins make through other host proteins. Nodes representing effectors are aligned in the top two rows: green nodes for P. syringae (Psy) effectors and purple nodes for effectors from the biotrophic oomycete, Hpa. Red nodes represent 30 NLRs included in the PPIN-1, and turquoise nodes represent Arabidopsis proteins showing interactions with NLRs. Gray edges represent protein-protein interactions assayed by yeast two-hybrid system in PPIN-1. The number after Hub indicates the number of interaction partners of the host protein in the main interactome AI-1 (Mukhtar et al., 2011). A single NLR can be connected with multiple effector targets, while an effector target can be connected to multiple NLRs. Only two NLRs show direct interactions with effector proteins (bold edges), while most other NLRs in the main network make indirect connections to effectors through host proteins. R gene activities are highly interconnected both with one another and with other immune proteins. This interconnectedness makes optimizing the immune response more difficult, as changes in one protein have a high probability of affecting the activity of another immune protein. The interactome network analysis was inferred from yeast two-hybrid associations.