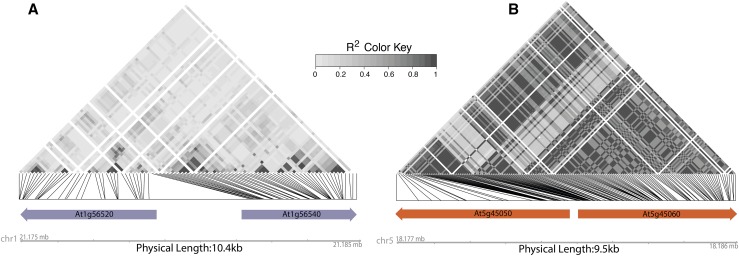
Figure 3.
Linkage between R Gene Haplotypes May Provide Evidence of Functional Cooperation and Coevolution.
Linkage across 762 Arabidopsis genotypes (Kawakatsu, 2016) is represented as R2 values of SNPs (minor allele frequency > 0.2) within R genes and visualized as a heat map. Only R genes are marked with arrows in these regions. Linkage disequilibrium in the region carrying At1g56520 and At1g56540 (A) is relatively low compared with that in the region carrying At5g45050 (RRS1B) and At5g45060 (RPS4B) (B). The increased linkage (higher R2) across the entire locus in (B) may be the result of the coevolution of alleles of the two R genes for their cooperative function. Functional cooperation of At5g45050 and At5g45060 (B) as a pair was demonstrated for disease resistance specificity as well as for tight cross-regulation against constitutive activity (Narusaka et al., 2009; Saucet et al., 2015). No such interaction was observed for the two R genes depicted in (A). If two R genes nearby or at a distance function together, mismatched allelic combinations could result in fitness costs. It is therefore reasonable to expect such R gene pairs to evolve tight linkage at either short range or long distance. Measuring linkage and population genomic statistics of R gene pairs in large sequencing projects, as we demonstrate here, may allow for the identification of candidate loci for interacting/coevolving pairs and allelic variants.