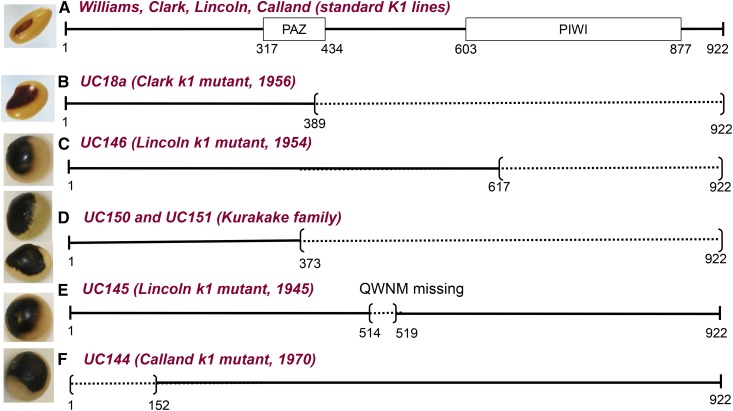
Figure 5.
Schematic of the Predicted Lesions in the AGO5 Protein Found in Independent Saddle Pattern Mutants.
(A) The AGO5 structure found in standard K1 lines and parents including Williams, Clark, Lincoln, and Calland showing the relative positions of PAZ and PIWI domains.
(B) Protein prematurely terminated in the k1 allele of the Clark 18a mutation.
(C) Protein prematurely terminated in the k1 allele of a 1954 saddle mutation in Lincoln.
(D) Proteins prematurely terminated in two different Kurakake lines, large seeded Plant Introduction lines collected in Japan.
(E) Protein missing four amino acids in a 1945 saddle pattern mutant of Lincoln.
(F) Protein of the saddle mutant found in Calland in 1970 and putatively missing a large part of its N terminus due to absence of the initiation codon.
Example seed phenotypes are illustrated. Dark lines represent amino acids of the AGO5 sequence, a tick mark indicates the relative position of altered amino acids introduced after a frameshift mutation, and dotted lines represent missing portions of the protein sequence. The Clark parent line is based on whole-genome resequencing data but the other nine are based on full length sequencing of the AGO5 amplicon. See Supplemental Data Set 5 for a more detailed characterization of the affected positions in the genes and proteins. Amino acid numbers shown underneath are relative to the initiation codon of Glyma.11G190900 in the standard K1 allele. All 10 lines sequenced showed an extra A in exon 20, which would increase the reading frame of the AGO5 protein to 922 amino acids rather than 890 as called by Phytozome for the Glyma.11G190900 predicted protein in the reference genome, representing a correction to the reference.