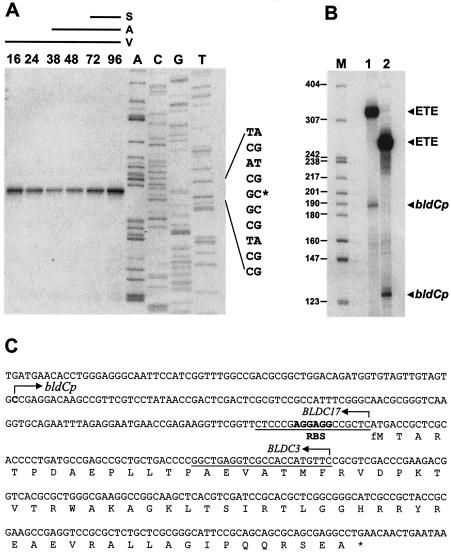
FIG. 8.
Transcriptional analysis of bldC. (A) High-resolution mapping of the 5′ end of the bldC message and analysis of bldC transcription levels during development of S. coelicolor M145 on R2YE solid medium. The time points in hours at which mycelium was harvested for RNA isolation and the presence of vegetative mycelium (V), aerial mycelium (A), and spores (S) are shown, as judged by microscopic examination. Lanes labeled A, C, G, and T represent a dideoxy sequencing ladder generated with the same radiolabeled oligonucleotide that was used to make the S1 mapping probe. (B) In vitro transcription of the bldCp promoter by S. coelicolor RNA polymerase holoenzyme. Transcripts were generated from template 1 (327 bp) or template 2 (264 bp) (see Materials and Methods). The expected sizes of the runoff transcripts from the bldCp promoter were 188 nucleotides (template 1) and 125 nucleotides (template 2). The size markers (M) were a 32P-end-labeled HpaII digest of pBR322. ETE, end-to-end transcription of the linear template. (C) Nucleotide sequence of the bldC gene showing the bldCp transcription start point, the putative ribosome-binding site (RBS), the complete bldC encoding sequence, and the sequences of the BLDC3 and BLDC17 downstream oligonucleotide PCR primers used to generate the templates.