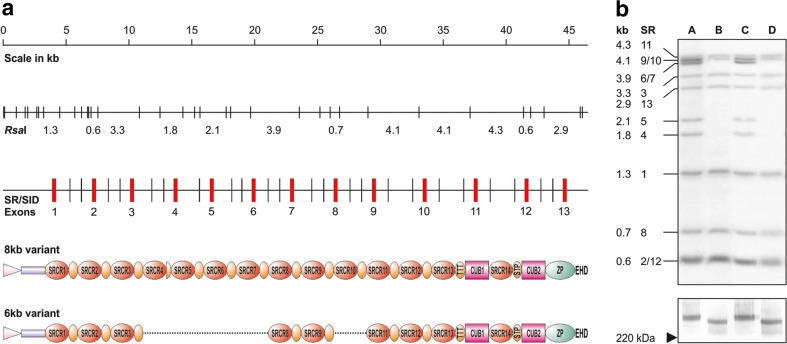
Fig. 1.
DMBT1 polymorphism leads to different length DMBT1 polypeptides. a Schematical presentation of the exon-intron structure within the relevant region of DMBT1 with resulting RsaI restriction fragment sizes depicted below. Gray boxes denote restriction fragments hybridizing with the probe DMBT1/sr1sid2. SR exons coding for scavenger receptor cysteine-rich domains. The hypothetical configurations within the proteins are depicted below. In the carboxy-terminal part of the protein resulting from the deleted allele, it cannot be discerned between a loss of either SR9, SR10, or SR11. Only one of the possibilities is shown. Pink triangle signal peptide, blue box motif without homology, SRCR scavenger receptor cysteine-rich domain, CUB C1r/C1s-Uegf-Bmp1 domains, ZP zona pellucida domain. EHD Ebnerin-homologous domain, orange ovals SRCR interspersed domains (SIDs), TTT and STP are threonine and serine-threonine-proline-rich domains, respectively. b Top panel Southern blot analysis of the DMBT1 genomic configuration in four individuals (A–D) selected from the panel. Band sizes and exons locating on the restriction fragments are depicted at the left. Bottom panel Western blot analysis of DMBT1SAG protein sizes in the partially purified and concentration-adjusted saliva samples of the four probands. The arrowhead denotes the position of the 220-kDa marker band. DMBT1SAG was collected from saliva donors that were homozygous for DMBT1/8 kb (donors A and C), homozygous for DMBT1/6 kb (donors B and D). Crude DMBT1SAG from the four donors samples were separated on 7.5% polyacrylamide gels, transferred to nitrocellulose and immunoblotted with mAb DMBT1H12 . Lane 1 donor A, lane 2, donor B; lane 3, donor C; lane 4, donor D. DMBT1SAG of donors A and C migrated at a position corresponding to an apparent molecular mass of approximately 340 kDa. DMBT1SAG of donors B and D runs at a position corresponding to approximately 255 kDa