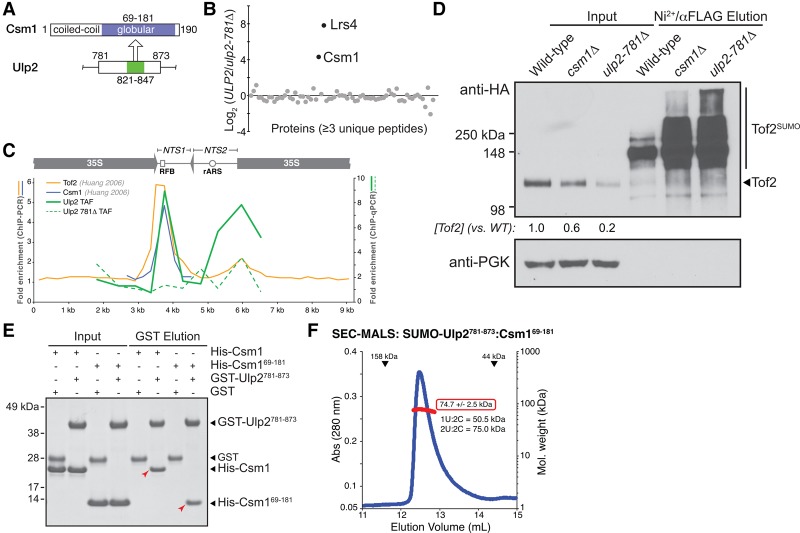
Figure 2.
Ulp2 binds the Csm1 C-terminal domain. (A) Schematic of the Ulp2-binding (residues 821–847; green) and Csm1-binding (residues 69–181; blue) regions. (B) Quantitative MS comparison of Ulp2-TAF-associated proteins (Chen et al. 2007) in ULP2-TAF versus ulp2-781Δ-TAF strains. Csm1 and Lrs4 are the only two proteins showing a strong enrichment in wild-type ULP2 cells compared with ulp2-781Δ cells. (C) ChIP-qPCR assay to detect the localization of Ulp2-TAF (solid green line) or Ulp2-781Δ-TAF (dashed green line) to rDNA repeats (average of triplicate samples shown, showing fold enrichment over untagged control). Localizations of Tof2 (orange) and Csm1 (blue) were measured previously by ChIP-PCR, reproduced here for comparison (Huang et al. 2006). (D) Western blot showing that the csm1Δ and ulp2-781Δ mutants have similar effects on Tof2 sumoylation and abundance. (E) GST pull-down assay showing a direct interaction between Csm1 (full-length or isolated C-terminal domain residues 69–181) and GST-Ulp2781–873. The Ulp2–Csm1 interaction is not affected by mutations to the conserved hydrophobic surface on Csm1 that has been implicated in binding Dsn1 and Tof2 (Supplemental Fig. S2A; Corbett et al. 2010). (F) Size exclusion chromatography/multiangle light scattering (SEC-MALS) analysis of SUMO-Ulp2781–873:Csm169–181. Elution volumes of 158- and 44-kDa standards are shown at the top. Calculated molecular weight (red box) indicates a stoichiometry of two Ulp2:two Csm1.