Figure 3.
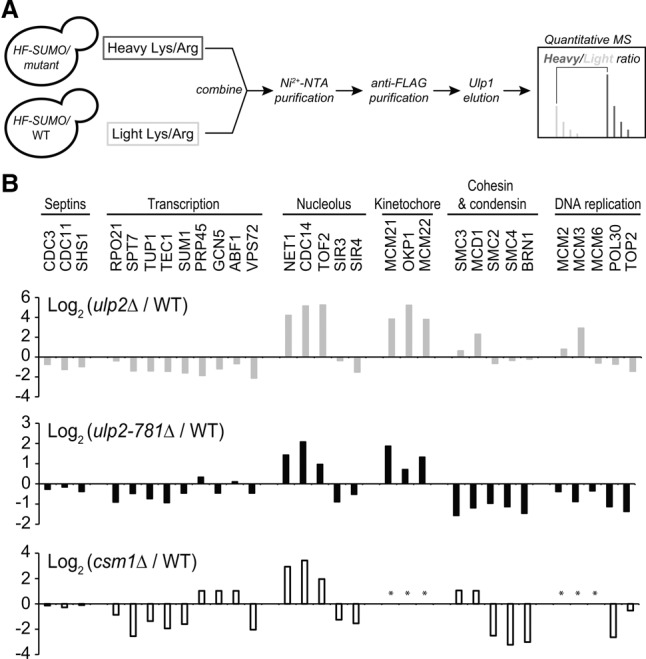
The Ulp2 C-terminal domain and Csm1 contribute to desumoylation of nucleolar and kinetochore proteins. (A) Experimental scheme for SILAC-MS to quantify differential sumoylation in ulp2-781Δ versus wild-type using HIS6-3xFlag-SMT3 (HF-SUMO). (B) Quantitative MS analysis. (Top panel) ulp2Δ versus wild type (gray bars) (de Albuquerque et al. 2016). (Middle panel) ulp2-781Δ versus wild type (black bars). (Bottom panel) csm1Δ versus wild type (black-outlined bars). Positive log2 values indicate proteins whose sumoylation increases in the mutant. Shown is a subset of proteins previously shown by other studies to be sumoylated and/or to be Ulp2 substrates (see Supplemental Tables 2, 3 for a complete list of the modified proteins identified). The asterisk indicates protein not found with at least three unique peptides.
