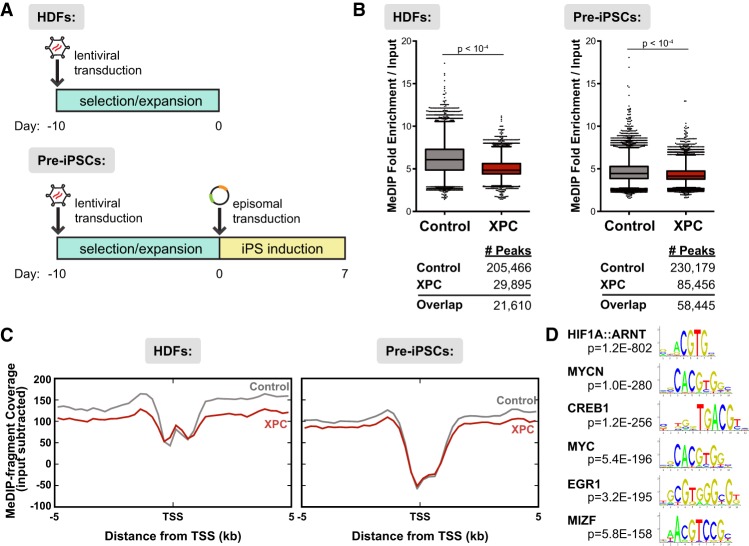
Figure 3.
Ectopic expression of XPC results in loss of DNA methylation genome-wide in both HDFs and pre-iPSCs. (A) Design of MeDIP-seq experiments. Reprogramming factors are cotransfected with a GFP-expressing plasmid to allow for sorting of transfected cells. (B) Up-regulation of XPC expression results in lower enrichment over background and fewer peaks called in MeDIP-seq analyses for both uninduced HDFs (left) and pre-iPSCs (right; 7 d post-induction). The number of peaks identified for each data set using MACS2 is shown below the graphs. (C) Distribution of MeDIP-seq reads by their distance ±5 kb from the TSSs of RefSeq genes, input subtracted. (D) Motif discovery of pre-iPSC MeDIP peaks enriched in the control but not the XPC gain-of-function data set.