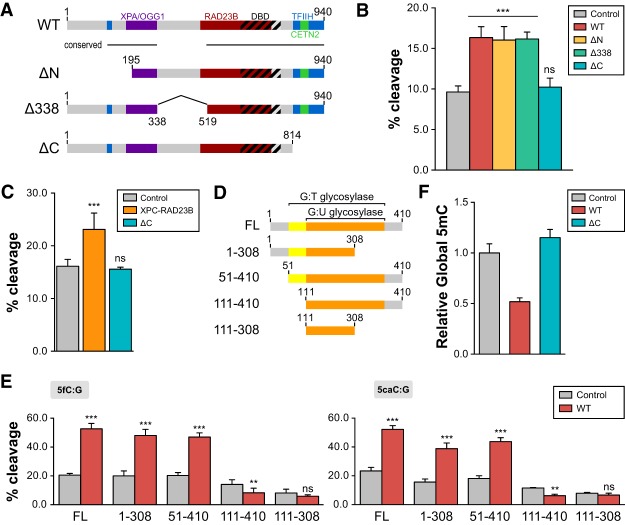
Figure 5.
XPC-mediated stimulation of TDG activity requires the C terminus of XPC and N terminus of TDG. (A) Schematic representation of human XPC (wild-type [WT]) highlighting the DNA-binding domain (DBD) and various protein–protein interaction domains. XPC truncations used in this study—ΔN (195–940 amino acids), Δ338 (del338–519 amino acids), and ΔC (1–814 amino acids)—are also shown. (B) TDG cleavage activity of a 5′-labeled T:G mismatch-containing oligonucleotide dsDNA in the presence of wild-type XPC or various XPC truncation complexes (ΔN, Δ338, and ΔC). (C) The CETN2 subunit is dispensable for stimulation of TDG-mediated base excision. TDG cleavage activity of a 5′-labeled T:G mismatch DNA substrate in the presence of the XPC–RAD23B heterodimer or a heterodimer containing ΔC XPC and RAD23B. (D) Schematic representation of full-length human TDG (FL) and truncations used in the in vitro glycosylase assay. The domains shown previously to be essential for G:U and G:T mismatch repair are indicated (Gallinari and Jiricny 1996). (E) Glycosylase activity of TDG truncations in the presence or absence of wild-type XPC was assayed using 5fC:G (left) and 5caC:G (right) substrates. The concentrations used were 0.2 µM DNA, 0.4 µM XPC complexes, and 50 nM TDG. (F) Relative global DNA methylation assayed by 5mC-specific ELISA using genomic DNA purified from HDFs overexpressing wild-type or ΔC human XPC. Error bars represent the standard deviation. n = 3. (***) P < 0.001; (**) P < 0.01; (n.s.) nonsignificant, calculated by two-way ANOVA.