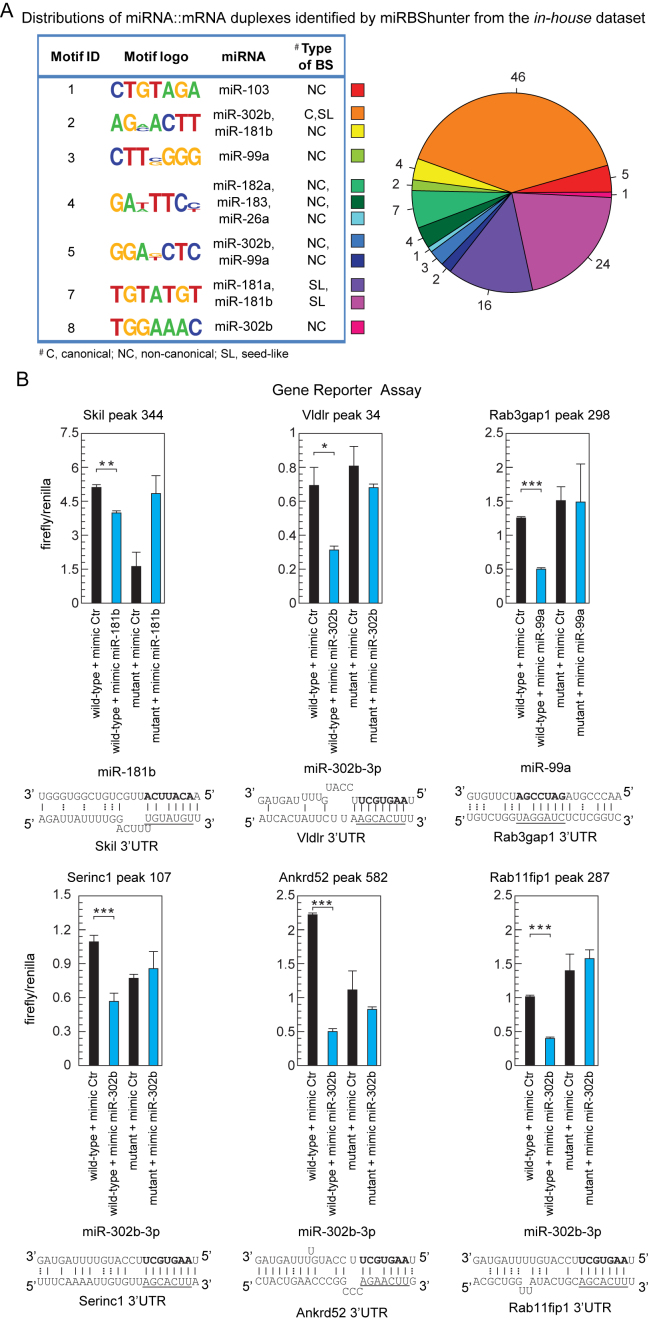
Figure 7.
Experimental validation of the miRNA binding sites found by our method. (A) Left panel: the motifs identified by miRBShunter for the in-house dataset with CS ≥ 1 are reported with the corresponding miRNA matching and the type of binding site (BS). Right panel: distribution of the different types of miRNA:mRNA duplexes identified by miRBShunter. Color code is indicated. (B) Relative luciferase activity of reporter constructs containing two either wild-type or mutant miRNA binding sites. HEK-293T cells were transfected with either the indicated mimic miRNA or control. The data were normalized using Renilla activity. miRNA::RNA heteroduplex structure expected to result from base pairing of the indicated miRNAs with the indicated binding sites are shown below the corresponding luciferase activity. A-U and G-C base pairs are represented by solid lines; G-U wobble base pairs are represented by dotted lines. Underlined the motif found by Homer software and matching with the portion of 7 nt of miRNA sequence in bold. Data are presented as mean and s.d. (n = 3). Student's t-test: *P < 0.05; **P < 0.01;; ***P < 0.001.