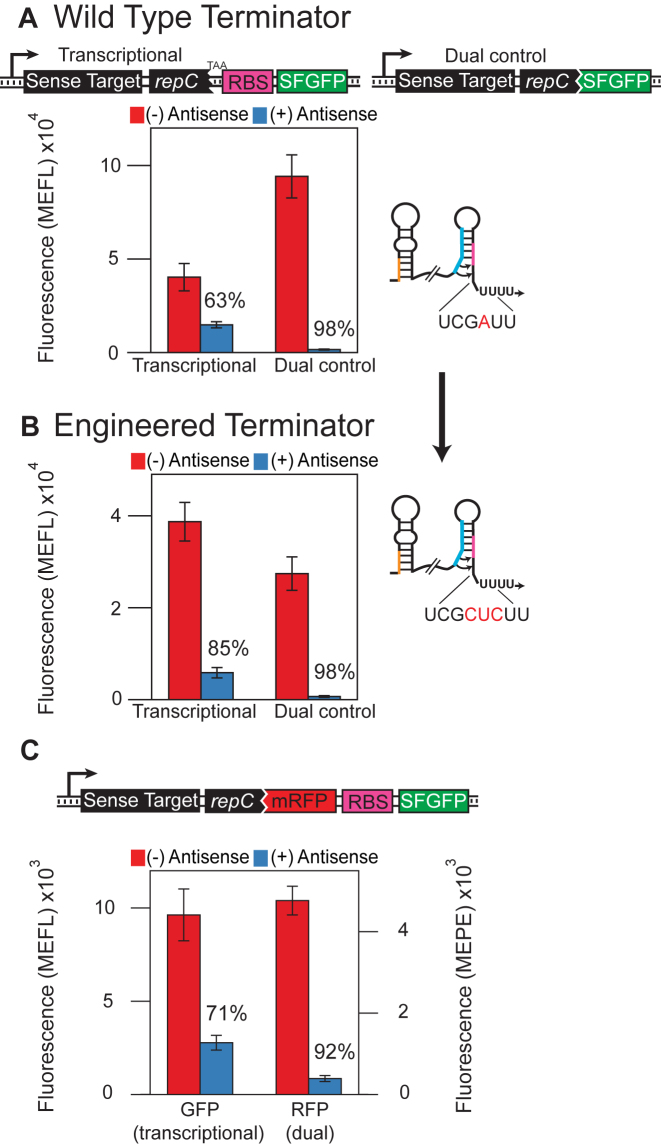
Figure 2.
Dual transcription/translation control represses gene expression with higher dynamic range than transcription control in vivo. Functional characterization of the (A) wild type (28), or (B) engineered (2) attenuator configured to repress either transcription (transcriptional fusion) or dual transcription/translation (translational fusion) of an SFGFP coding sequence. Average fluorescence was collected by flow cytometry as Molecules of Equivalent Fluorescein (MEFL) of E. coli TG1 cells transformed with a plasmid expressing the indicated attenuator-SFGFP construct and a plasmid expressing the antisense RNA (+, blue) or a control plasmid lacking the antisense sequence (–, red) (Supplementary Table S2). Percent repression is labelled above each construct tested. In both cases the dual control regulator showed 98% repression (50-fold), though with a higher ON expression level for the wild type attenuator. Error bars represent standard deviations of at least seven biological replicates. Cartoons highlight differences between the wild type and engineered attenuator sequences, which differ by several bases in the 3΄ half of the terminator hairpins (Supplementary Figure S8). (C) Testing dual control versus transcriptional control in a two-color operon construct. The wild type attenuator sequence was translationally fused to an mRFP coding sequence, which was followed by an RBS-SFGFP sequence. In this way, mRFP was under dual transcription/translation control while SFGFP was under only transcription control. The construct was tested as in (A) with mRFP fluorescence collected by flow cytometry as Molecules of Equivalent Phycoerythrin (MEPE). RFP was more strongly repressed at 92% (±1.7%) than GFP at 71% (±5.8%). Averages and standard deviations plotted in (A) and (B) are presented Supplementary Table S3 to allow for comparison within orders of magnitude.