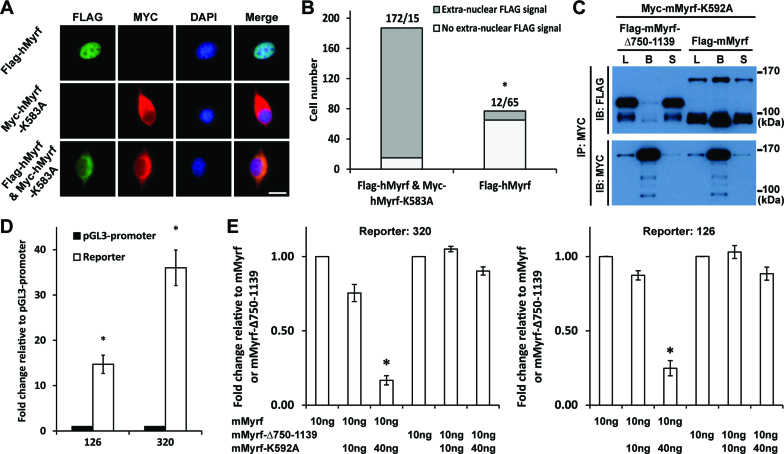
Figure 3.
Myrf N-terminal fragments form homo-trimers prior to ER release. (A) Flag-hMyrf and Myc-hMyrf-K583A were transfected into CG4 cells alone or together to determine the subcellular localization of their N terminal fragments. Scale bar, 10 μm. (B) The immunofluorescence result in panel A was quantified by cell counting. The binomial test showed that the N-terminal fragments of Flag-hMyrf were found outside the nucleus more frequently in the presence of co-expressed Myc-hMyrf-K583A (P value < 5.72 × 10−119). (C) Flag-mMyrf and Myc-mMyrf-K592A were co-transfected into HEK293FT cells. Cell lysate (‘L’) was subject to immunoprecipitation with Myc beads, leading to the bound (‘B’) and sup (‘S’) fractions. Flag-mMyrf-Δ750-1139 was used for a control immunoprecipitation experiment. (D) Two luciferase reporters (126 and 320), chosen from the published Myrf ChIP-seq data (14), responded well to transfected Myrf. The response of pGL3-promoter was set to 1, and the reported values are means and standard errors of 3 biological replicates. *P < 0.05 by two-tailed one sample Student's t test with Bonferroni correction for multiple hypothesis testing. (E) Luciferase assay determining how the co-expression of mMyrf-K592A affects the transcriptional activity of mMyrf or mMyrf-Δ750-1139. The transcriptional activity of mMyrf was set to 1, and the reported values are means and standard errors of 4 biological replicates. Two-way ANOVA showed that there was a significant interaction between the type of mMyrf (mMyrf or mMyrf-Δ750-1139) and the amount of co-expressed mMyrf-K592A. *P < 7.5 × 10−6 for 320, and P < 0.01 for 126. Post-hoc analysis indicated that mMyrf-K592A significantly suppressed the transcriptional activity of mMyrf, but not mMyrf-Δ750–1139. *P < 0.0003 for 320 and P < 0.02 for 126. IP: immunoprecipitation. IB: immunoblotting.